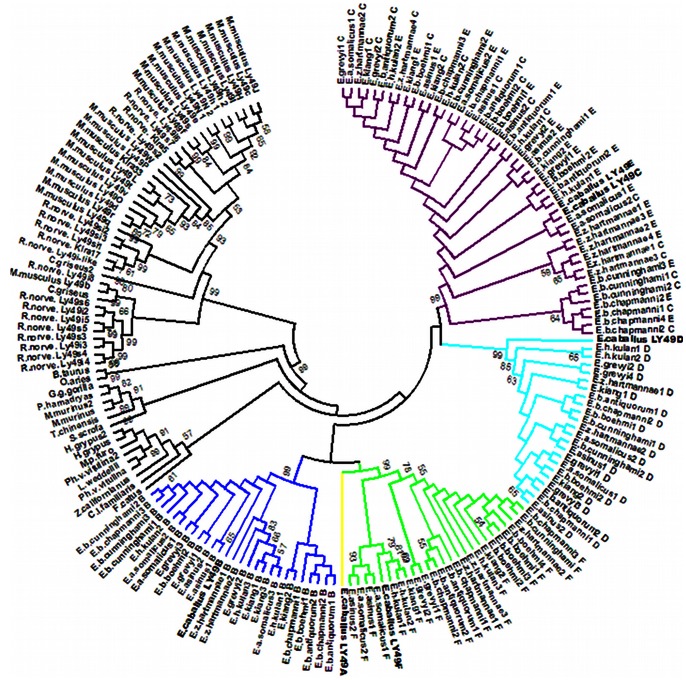
Figure 1. Phylogenetic tree of mammalian Ly49 C-type lectin-like domain sequences.
The evolutionary history was inferred using the neighbor-joining method [39]. The bootstrap consensus tree inferred from 1000 replicates [40] is taken to represent the evolutionary history of the taxa analyzed: Mus musculus (NP_032489.1, NP_032490.1, NP_444384.1, NP_034781.2, NP_034778.2, NP_032485.2, NP_444381.1, NP_001095090.1, NP_032487.2, NP_034780.1, NP_001239078.1, NP_038821.2, NP_444382.1, NP_444380.1, NP_034776.1, NP_077790.1, NP_444383.1, NP_001034207.1, NP_001239506.1, NP_573466.3, NP_038822.3, NP_032488.4); Rattus norvegicus (NP_001009718.1, NP_775413.1, NP_942041.1, NP_001009494.1, NP_001009919.1, NP_001009497.1, NP_001009498.1, NP_001104780.1, NP_001165559.1, NP_001009495.1, NP_001009499.1, NP_714948.1, NP_001012767.1, NP_001009487.1, NP_690061.1, NP_001009501.1, NP_001009486.1, NP_001009488.1); Cricetulus griseus (XP_003510604.1, XP_003509313.1); Bos taurus (NP_776801.1); Ovis aries (XP_004006913.1); Gorilla gorilla gorilla (XP_004052762.1); Papio hamadryas (AAK26161.1); Microcebus murinus (ACO83129.1, ACO83128.1); Tupaia chinensis (ELV12449.1); Sus scrofa (AAP13541.1); Halichoerus grypus (ACN78613.1, ACN78614.1); Mustela putorius furo (AES00881.1); Phoca vitulina vitulina (ACN78615.1, ACN78616.1); Leptonychotes weddellii (ACN78617.1); Zalophus californianus (ACN78618.1); Canis lupus familiaris (AAP13540.1); Felis catus (AAP13539.1); Equus caballus (NP_001075297.1, NP_001075298.1, NP_001075299.1, NP_001075392.1, NP_001075393.1, NP_001075998.1); Equus grevyi; Equus zebra hartmannae; Equus burchellii boehmi; Equus burchellii antiquorum; Equus burchellii chapmanni; Equus burchellii cunninghami; Equus asinus; Equus asinus somalicus; Equus kiang and Equus hemionus kulan. Bootstrap confidences over 50% are given as numbers. The evolutionary distances were computed using the p-distance method [41]. The analysis involved 184 amino acid sequences. All ambiguous positions were removed for each sequence pair. A total of 135 positions were included into the final dataset. Evolutionary analyses were conducted in MEGA5 [26].