Abstract
The measurement of the volume of the human amygdala in vivo has received increasing attention over the past decade, but existing methods face several challenges. First, due to the amorphous appearance of the amygdala and the difficulties in interpreting its boundaries, it is common for protocols to omit sizable sections of the rostral and dorsal regions of the amygdala comprising parts of the basolateral complex (BL) and central nucleus (Ce), respectively. Second, segmentation of the amgydaloid complex into separate subdivisions is challenging due to the resolution of routinely acquired images and the lack of standard protocols. Recent advances in technology have made ultra-high resolution MR images available, and in this study we provide a detailed segmentation protocol for manually tracing the whole amygdala that incorporates a greater portion of the rostral and dorsal sections with techniques illustrated in detail to maximize reproducibility. In addition, we propose a geometrically-based protocol for segmenting the amygdala into four component subregions of interest (sROI), which correspond largely to amygdala subnuclear divisions: the BL sROI, centromedial (CM) sROI, basomedial (BM) sROI, and the amygdaloid cortical (ACo) sROI. We performed an intra- and inter-rater reliability study of our methods in 10 adults (5 young adults and 5 older adults). The results indicate that both protocols can be implemented with a high degree of reliability (the majority of intra-rater and inter-rater correlations were >0.81). This protocol should aid further research into the alterations in amygdala anatomy, connectivity, and function that accompany normal aging and pathology associated with neuropsychiatric disorders.
Keywords: Amygdala, Segmentation, MRI, Subnuclei, Parcellation
Introduction
The amygdala is an amorphous gray matter structure within the rostral medial temporal lobe. It has been implicated in many psychological phenomena, including emotion and affect (e.g. Adolphs et al., 1994; Barrett et al., 2007; Lanteaume et al., 2007; LeDoux et al., 1988; Sharot et al., 2007), social behavior (e.g. Amaral, 2003; Machado et al., 2008; Rosvold et al., 1954), attention (e.g. Pessoa et al., 2002a, 2002b; Ursin and Kaada, 1960), perception (e.g. Sander and Scheich, 2001; Whalen et al., 1998), learning (e.g. Gaffan et al., 1989; Hooker et al., 2006; Morris et al., 1998), and memory (e.g.Fadok et al., 2010; Packard et al., 1994). This is not surprising given its dense connections with many regions of the brain (for a review, see Swanson and Petrovich, 1998).
In living humans, investigations of amygdala function and dysfunction have primarily employed structural and functional neuroimaging tools (in addition to studies of patients with lesions). Although a number of studies have been performed on the relationship of amygdala volume to specific behaviors and in aging and neuropsychiatric disorders (for reviews, see Anand and Shekhar, 2003; Phelps and LeDoux, 2005; Wright, 2009), heterogeneous results are present likely in part because of the variety of approaches to volumetric measurement of the amygdala.
Over the past 20 years, a number of manual tracing protocols have been published that provide methods for measuring human amygdalar volume (Achten et al., 1998; Bonilha et al., 2004; Convit et al., 1999; Makris et al., 1999; Matsuoka et al., 2003; Pruessner et al., 2000; Watson et al., 1992). Although these approaches have been very useful for identifying a variety of interesting effects, we believe that they and automated segmentation protocols based on similar manual tracings (e.g., Freesurfer’s automatic segmentation (Fischl et al., 2002)) can be improved in a number of ways. First, the method for demarcating the dorsal boundary of the caudal amygdala is not clearly described or illustrated, making reproduction of published techniques challenging (Achten et al., 1998; Convit et al., 1999; Watson et al., 1992). The borders of this region are typically difficult to see in structural MR imaging so a heuristic conservative approach may be employed to interpolate a line along the ventral edge of the optic tract. However, this means that the central nucleus (Ce)–a key visceromotor regulation site that is considered to be a major “output” nucleus of the amygdala–is often omitted (Price and Amaral, 1981). Similarly, the location of the anterior boundary of the amygdala is not clearly described or illustrated in prior protocols. Finally, the resolution of the images on which these prior protocols were based (typically about 1 mm isotropic voxels) is a limiting factor, since at this resolution it is difficult to see much of the fine structure of this region, including gray-white boundaries around the perimeter of the amygdala.
Next, although there have been several prior attempts to subdivide the amygdala into component parts (including the divisions into ventral vs. dorsal amygdala (Davis et al., 2010; Whalen et al., 2001)), there is no published protocol for the manual segmentation of amygdala subnuclei using structural MRI. The measurement of amygdala subnuclear volume is of potential value given distinct functional roles proposed for these regions. Although debate continues about the subdivisions of the amygdala at the histological level (Swanson and Petrovich, 1998), one basic approach is to consider a macro-level segmentation into the basolateral complex (BL), centromedial complex (encompassing the Ce and medial nucleus), basomedial complex (BM), and the amygdaloid cortical complex (ACo) (Paxinos and Mai, 2004).
Amunts and colleagues published the first study to match histological and MRI data in an attempt to develop a histologically-based probabilistic atlas of subdivisions of the amygdala (Amunts et al., 2005). Their protocol involved generating 3D reconstructions of digital photomicrographs of the histological samples from 10 post-mortem brains and manually tracing three subdivisions (the CM group, the BL group, and the superficial group) directly onto the photomicrograph images. The ten 3D-reconstructed images were registered to MNI template space to produce probabilistic maps of the amygdala subregions. The application of this protocol necessitates manipulation of individual subjects’ MRIs into standard template (MNI) space, a process that can produce inaccuracies due to the deformations required (Yassa and Stark, 2009). Moreover, the probabilistic nature of the method requires that the user measures subnuclear volume in areas in which voxels of different subnuclei do not overlap (e.g., > 50–75% probability), which underestimates the full extent of the volume of each subnuclear region in any individual subject.
Newer methods using diffusion tensor imaging (DTI) to segment the amygdala have added a perspective from connectional anatomy to parcellation endeavors (Bach et al., 2011; Saygin et al., in press; Solano-Castiella et al., 2010). Solano-Castiella et al. (2010) identified two primary diffusion directions of amygdala voxels and used this information to separate the amygdala into medial and lateral subdivisions. Further work has shown that diffusion-based segmentations correspond to anatomical connectivity (Bach et al., 2011). One challenge in using DTI for amygdala subnuclear segmentation is its low spatial resolution (although the data in Solano-Castiella et al., 2010 is relatively high resolution for DTI, at 1.7 mm in-plane and 1.7 mm thickness). Similar resolution issues exist for resting-state fMRI-based connectivity analysis (3 mm slice thickness in Roy et al., 2009).
Here we used the clarity of ultra high-resolution scans to create a detailed approach to the segmentation of the amygdala using a widely available histological atlas as our guide. Such high-resolution MR images have not been used before for amygdala segmentation, although they have been used previously in a study of hippocampal subregions (Van Leemput et al., 2009). These images provide a much higher level of visible detail than is typically present in routine 1 mm scans. We used ultra-high resolution images from five young subjects and five cognitively normal elderly subjects to trace both the whole amygdala and its major subregions in living humans. Our first step was to create a new protocol for capturing more of the dorsal and anterior portions of the whole amygdala. Next, we segmented the whole amygdala into four separate geometrically-defined subregions. It is important to point out that these geometrically-defined regions do not correspond exactly to specific amygdalar subnuclei, but rather provide subregions of interest that approximate the major subdivisions of the amygdala. We have chosen to use the following terms for these subregions: basolateral subregion of interest (BL sROI), basomedial sROI (BM sROI), centromedial sROI (CM sROI), and amygdaloid cortical sROI (ACo sROI).
After all tracings were completed, we examined the reproducibility of these operations by comparing the intra- and inter-rater reliability of manual tracings of both the whole amygdala and its subregions done by two operators as a function of voxel overlap and intraclass correlation of volumes. Finally, we provide here a detailed set of images from our own MRI data and also corresponding images reproduced from the Mai et al. (1997) atlas to assist in the reproduction of this method or the comparison of this approach to others.
Methods
Participant information
For these experiments, we used scans of the brains of 10 human subjects (5 younger individuals, 4/1 males/females, age range 22–25, mean/s.d.=23.8/1.3; and 5 older cognitively normal individuals, 3/2 males/females, age range 72 to 78; mean/s.d.=74.8/2.6). All younger subjects were screened and underwent a structured interview to determine that they had no history of neurologic or psychiatric disorders, including head injury or substance abuse. All older subjects were evaluated clinically through structured interviews and neurologic and cognitive examination and determined to be cognitively normal and without history of neurologic or psychiatric disorders, including head injury or substance abuse.
MRI scan acquisition
MRI scan data were collected using a prototype custom-built 32-channel head coil (Wiggins et al., 2006) with 3.0 Tesla Siemens Total Imaging Matrix Trio system (Siemens Medical Systems, Iselin, NJ). T1-weighted data were acquired using an anisotropic MPRAGE sequence obtained in an oblique coronal orientation (perpendicular to the long axis of the hippocampal formation) with 380 μm in-plane resolution with the following parameters: TR//TI//TE=2530//1100//5.39 ms, FOV=170 mm, matrix=448, FA=7°, 208 slices acquired coronally, thickness=0.8 mm, GRAPPA acceleration factor=2, acquisition time=7.34 min. Five separate acquisitions were collected and averaged together with 6-parameter affine registration to obtain a single high signal-to-noise ratio volume.
Whole amygdala segmentation protocol
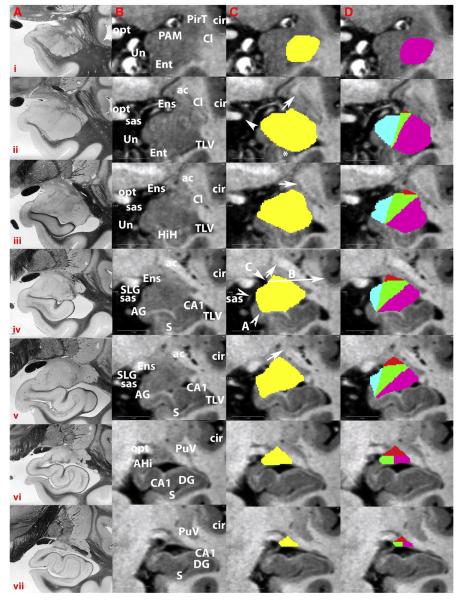
Two independent raters used the Freeview tool in the Freesurfer software suite (Massachusetts General Hospital, Boston, MA; Dale et al., 1999; Fischl et al., 1999a, 1999b, 2001; Fischl and Dale, 2000; Ségonne et al., 2004) (http://surfer.nmr.mgh.harvard.edu/) to perform the manual tracing. Tracing began in the coronal plane starting with the first slice rostral to the rostral tip of the hippocampal formation (Fig. 1Cii, asterisk). From this initial slice located in the mid-portion of the amygdala, the amygdala was traced in sequential slices moving to its most rostral border and then backward to its most caudal border (Fig. 1). The rostral boundary of the amygdala is difficult to identify given the gradual transition from cortex, but the high resolution of the images allowed both raters to identify the same rostral boundary (same coronal slice) based on visual inspection; this is the most rostral slice on which nuclear gray matter could be seen subjacent to the cortex (see Results). The caudal boundary of the amygdala was similarly identified using a purely visual method. Moving caudally, the whole amygdala was traced until it could no longer be seen as a distinct nucleus after it ascended and became impossible to discern from the ventral striatum, nucleus basalis, and stria terminalis (see Results for reliability analysis of this boundary).
Fig. 1.
Comparison of histological photographs from Mai atlas (Mai et al., 1997) with MR images (380 μm in-plane voxel size) and tracings on one young subject at different levels, from most rostral (row i) to most caudal (row vii) boundaries. Columns show A) histological images; B) MRI slices; C) whole amygdala tracings in yellow; D) amygdalar subregion tracings following the protocol illustrated in idealized terms in the next figure: amygdaloid cortical complex (ACo sROI) is blue; basolateral complex (BL sROI) is purple; basomedial complex (BM sROI) is green; centromedial complex (CM sROI) is red. Arrows and arrowheads illustrate specific points described in the Methods section detailing the protocol. Post-mortem histological sections are shown to provide a standard set of reference images to illustrate the landmarks in our protocol. Note that there are multiple differences between the two types of images in contrast properties. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Images in column A are reproduced with permission from Elsevier Inc.
Rostrally, the dorsal boundary of the amygdala could be seen within the white matter ventral to the piriform cortex. Moving caudally, once the semilunar gyrus (SLG) appeared, the dorsomedial border became the cerebrospinal fluid (CSF). At this point, the dorsolateral edge was assigned by tracing a diagonal line from the most lateral point of the endorhinal sulcus to the most lateral point of the anterior commissure (AC) (Fig. 1Cii, solid arrow and Figs. 2.2, 2.3). As the amygdala extended caudally and the AC was positioned directly ventral to the putamen, we drew this line from the endorhinal sulcus to the midpoint of the AC (Fig. 1Ciii, solid arrow and Figs. 2.4, 2.5). Further caudally, the line was drawn to intersect the medial edge of the AC when it connected to the white matter of the temporal stem (Fig. 1Civ and 1Cv, solid arrow and Figs. 2.6, 2.7). When the AC was no longer visible, the boundary was traced along the white matter band wrapping around the dorsolateral amygdala (Fig. 1Cvi and Figs. 2.8, 2.9).
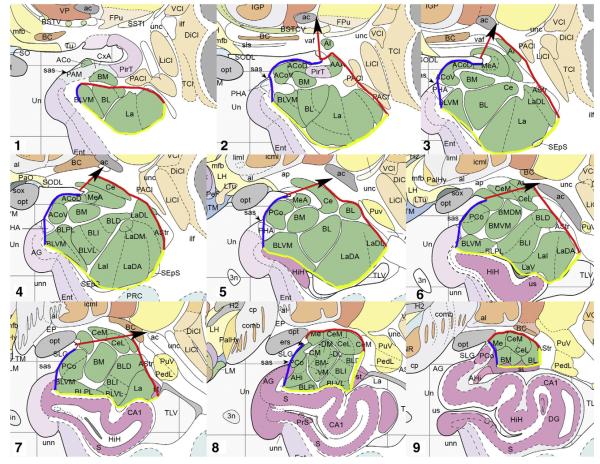
Fig. 2.
Illustration of idealized boundaries manually drawn on Mai atlas figures (Mai et al., 1997), from most rostral (1) to most caudal (9). Medial boundaries are in blue; dorsal and dorsolateral boundaries are in red; ventral and ventro-lateral boundaries are in yellow. Red arrows are shown to indicate direction of dorsal boundary toward anterior commissure; arrows are not part of the tracing protocol. See Box 1 for abbreviations. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Images are reproduced with permission from Elsevier Inc.
The dorsolateral and ventrolateral boundaries were the easiest to identify because the lateral edge of the amygdala is bordered by white matter and/or CSF from the temporal horn. These borders were extended until they connected with the dorsal and ventral boundaries, respectively.
The ventral boundary of the amygdala is initially white matter. Once the hippocampal formation appeared, the ventral boundary was established by tracing from the most ventral point of the amygdala along the white matter strand of the alveus toward the midpoint of the optic tract in the most rostral slices (Fig. 1Cii, arrowhead and Fig. 2.1). Once the semiannular sulcus (SAS) is visible, the ventromedial boundary was established by interpolating a line from the medial tip of the gray matter of the amygdala up to the fundus of the SAS (Figs. 2.2–2.4). Moving caudally as the hippocampus appears, the ventral boundary of the amygdala is traced along the alveus to its medial tip (Fig. 1Civ, lower arrowhead), and then a line is interpolated to the fundus of the SAS (Fig. 1Civ, upper arrowhead and Figs. 2.5–2.8). This approach is useful for precision at the possible expense of accuracy at the most rostral levels of amygdala, since it will include some piriform cortex in a few slices.
The medial boundary of the amygdala is mostly defined by its adjacency to CSF. However, on caudal slices the boundary was defined arbitrarily as the narrowest point along the isthmus of the hippocampal–amygdalar transition area (Fig. 1Cvi).
Amygdala subregion protocol
We independently traced the amygdalar subregions of all five young and five elderly subjects using the outlines of the whole amygdala tracings derived from one operator. The amygdala was separated into four subnuclear divisions: the BL sROI (including both the basolateral nucleus and the lateral nucleus), BM sROI, CM sROI (including both the central nucleus and the medial nucleus), and the ACo sROI (including all superficial amygdalar nuclei) (see Fig. 1).
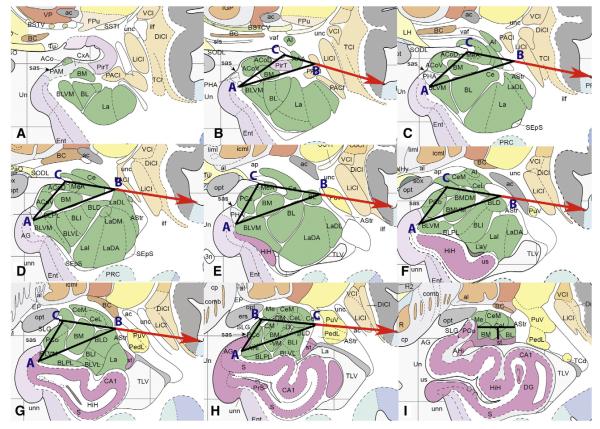
Beginning from the rostral boundary of the whole amygdala, the entire structure was labeled as BL sROI because the amygdala is primarily basolateral nucleus complex at this point. Although there is a small section of BM sROI present at this level, it was not possible to visualize (Fig. 3A). Once amygdalar tissue rose above the SAS, we began tracing the subregions according to a geometric method (Fig. 3B). The first step in this process was to identify three key points on the whole amygdala tracing. Point A was the medial tip of the alveus (Fig. 1Civ, lower arrowhead labeled A). Point C was the most lateral point of the endorhinal sulcus (Fig. 1Civ, upper arrowhead labeled C). Point B was determined by drawing a line from Point C to the bottom of the circular sulcus (Fig. 1Civ, long arrow); the point where this line intersected the dorsolateral boundary of the whole amygdala tracing became Point B (Fig. 1Civ, intersection between long arrow and yellow region of interest labeled B; see also Fig. 3). We chose these three points because they were easily observable and provided a reliable landmarking system for segmenting the amygdala along internally established axes.
Fig. 3.
Detail of geometrically determined protocol for segmenting the amygdala, from most rostral (A) to most caudal (I). See text for explanation of points A, B, and C. A is not segmented because the whole amygdala tracing is labeled as BL at this level. Horizontal line from Point C to Point B extends beyond boundaries of whole amygdala to indicate direction toward the circular sulcus, but ends at Point B in the protocol. I is split evenly on the horizontal axis and on the vertical axis below the level of the central nucleus. See Box 1 for abbreviations.
Images are reproduced with permission from Elsevier Inc.
After identifying these points, the next step was to draw straight lines connecting the three points to each other. These lines divide the whole amygdala tracing into 4 quadrants (Fig. 3). The four quadrants represent an approximation of the four subnuclear regions, including the ACo sROI (medial), BL sROI (lateral), CM sROI (dorsal), and BM sROI (central). We used this approach for the rostral portion of the protocol. Although some voxels from non-amygdala structures are present in our designation of BM sROI (e.g. piriform cortex (PirT)), we chose to be inclusive rather than to exclude a relatively large portion of the dorsal amygdala.
The caudal portion of the protocol began with the first slice in which the isthmus of the hippocampal–amygdalar transition area was visible with an open hippocampal sulcus (Fig. 3I). Beginning with this slice, the whole amygdala was then divided into three subregions: the CM sROI, the BM sROI, and the BL sROI. At this point, the amygdala is quite small and it is necessary to measure the distance, voxel by voxel, on the vertical and horizontal axes of the amygdala tracing. We visually divided the tracing into two halves using a horizontal line, labeling the top half the CM sROI; if the number of voxels was odd, we included the extra row of voxels in the CM sROI due to its greater size at this section of the amygdala. We further subdivided the bottom half of the whole amygdala tracing with a vertical line extending from the midpoint of the horizontal axis to the most ventral point of the whole amygdala tracing: the medial section was labeled as the BM sROI and the lateral section was labeled as the BL sROI (Fig. 3I). If there was an even number of voxels, we allotted the extra column of voxels to the BL sROI because it is usually larger than the BM sROI at this stage.
We calculated the total volume of the whole amygdala and subregions in each subset of tracings in voxels and mm3 using tools in the Freesurfer software package. In addition to examining absolute volumes of the structures, we also examined volumes adjusting for intracranial volume by dividing each amygdalar volumetric measure by total intracranial volume obtained from Freesurfer’s automated subcortical segmentation processing stream, and multiplying by 1000.
Reliability analysis
The operators independently traced the entirety of the whole amygdala using the protocol described above. To determine the rostral and caudal boundaries of the whole amygdala, two operators independently selected the first and last slices in which they believed the amygdala was visible. These selections were compared for reliability.
After tracing the whole amygdala, one set of tracings was chosen as a template to be superimposed over the temporal lobe of each subject. Both operators subsequently divided this template into the four subregions of the amygdala. This method was used to control for inter-rater variation in the whole amygdala tracings.
We calculated the Model 2 intraclass correlation (ICC) (Shrout and Fleiss, 1979) of volumetric measurements between the tracings of each structure by each operator using SPSS 16.0 (Chicago, IL). The intra-class correlation analysis tested the similarity in the absolute volumes of the whole amygdala and its subregions as determined by each operator’s tracings. We followed the evaluative criteria set forth by Landis and Koch (1977), in which agreement measures of correlation of 0.81–1.00 are ‘almost perfect’, 0.61–0.80 are ‘substantial’, and 0.41–0.60 are moderate.
As an additional test of reliability, we obtained the Dice coefficient of voxel overlap between these two operator’s tracings, which is defined as the number of voxels overlapping between two tracings divided by the mean number of voxels in the two tracings (Dice, 1945). This measure determines whether both operators were demarcating the same voxels regardless of size of the structure.
Results
Whole amygdala
The two operators chose the most rostral slice with a visible amygdala subjacent to the piriform cortex with an intraclass correlation (ICC) of 0.998 across the first five subjects. The operators designated the most caudal slice with visible amygdala with an ICC of 0.998, as well. Thus, the selection of the most rostral and most caudal slices was highly reliable. The volumetric data for the whole amygdala are presented in Table 1.
Table 1.
Mean absolute (abs) and relative (rel) volumes and standard deviations of the whole amygdala and amygdala subregions in the left and right hemispheres. All volumes are reported in mm3. Relative volumes were calculated by dividing each amygdalar volumetric measure by total intracranial volume and multiplying by 1000. BL sROI = basolateral subregion of interest; BM sROI = basomedial sROI; CM sROI = centromedial sROI; ACo sROI = amygdala corticoid sROI.
| Operator PD |
Operator JE |
Operator JE (round 2) |
||||
|---|---|---|---|---|---|---|
| L | R | L | R | L | R | |
| Whole amygdala | ||||||
| Mean abs. | 1697.60 | 1707.78 | 1752.22 | 1766.82 | 1733.06 | 1776.38 |
| SD | 160.38 | 197.82 | 219.95 | 242.86 | 153.40 | 216.10 |
| Mean rel. | 1.068 | 1.111 | 1.099 | 1.097 | 1.115 | 1.080 |
| SD | 0.137 | 0.277 | 0.109 | 0.220 | 0.188 | 0.266 |
| BL sROI | ||||||
| Mean abs. | 1200.78 | 1061.32 | 1093.94 | 1027.92 | 1187.32 | 1076.46 |
| SD | 128.66 | 86.42 | 117.06 | 79.07 | 117.88 | 68.27 |
| Mean rel. | 0.666 | 0.694 | 0.646 | 0.758 | 0.678 | 0.762 |
| SD | 0.089 | 0.161 | 0.096 | 0.207 | 0.110 | 0.180 |
| BM sROI | ||||||
| Mean abs. | 286.94 | 313.86 | 317.12 | 352.82 | 294.32 | 328.52 |
| SD | 44.62 | 69.62 | 39.02 | 64.99 | 59.78 | 77.56 |
| Mean rel. | 0.195 | 0.202 | 0.219 | 0.184 | 0.202 | 0.182 |
| SD | 0.038 | 0.055 | 0.033 | 0.037 | 0.026 | 0.045 |
| CM sROI | ||||||
| Mean abs. | 145.34 | 179.74 | 191.38 | 180.90 | 149.22 | 163.60 |
| SD | 39.10 | 46.95 | 58.32 | 61.98 | 44.87 | 51.64 |
| Mean rel. | 0.114 | 0.120 | 0.113 | 0.093 | 0.101 | 0.092 |
| SD | 0.038 | 0.038 | 0.037 | 0.026 | 0.027 | 0.030 |
| ACo sROI | ||||||
| Mean abs. | 125.84 | 153.24 | 149.86 | 146.04 | 121.88 | 141.84 |
| SD | 35.74 | 75.09 | 25.52 | 59.62 | 31.63 | 42.46 |
| Mean rel. | 0.093 | 0.096 | 0.090 | 0.078 | 0.088 | 0.082 |
| SD | 0.034 | 0.034 | 0.027 | 0.027 | 0.025 | 0.040 |
| Young subjects (N=5) | ||||||
| Whole amygdala | ||||||
| Mean abs. | 1615.38 | 1483.58 | 1554.80 | 1523.34 | 1516.44 | 1469.00 |
| SD | 249.85 | 225.81 | 177.46 | 221.37 | 303.93 | 206.06 |
| Mean rel. | 1.076 | 0.992 | 1.035 | 1.007 | 1.016 | 0.979 |
| SD | 0.160 | 0.161 | 0.148 | 0.155 | 0.184 | 0.170 |
| BL sROI | ||||||
| Mean abs. | 1014.08 | 897.34 | 1006.34 | 942.30 | 1056.28 | 999.46 |
| SD | 106.27 | 153.72 | 67.98 | 135.88 | 118.78 | 161.62 |
| Mean rel. | 0.675 | 0.607 | 0.672 | 0.633 | 0.703 | 0.673 |
| SD | 0.102 | 0.132 | 0.110 | 0.128 | 0.110 | 0.134 |
| BM sROI | ||||||
| Mean abs. | 232.14 | 312.98 | 280.42 | 342.56 | 269.28 | 310.80 |
| SD | 47.48 | 53.57 | 57.41 | 59.99 | 55.17 | 57.67 |
| Mean rel. | 0.209 | 0.185 | 0.228 | 0.180 | 0.208 | 0.155 |
| SD | 0.040 | 0.027 | 0.039 | 0.039 | 0.043 | 0.031 |
| CM sROI | ||||||
| Mean abs. | 147.38 | 156.02 | 178.26 | 132.04 | 160.38 | 117.24 |
| SD | 34.18 | 29.35 | 26.25 | 21.40 | 23.65 | 19.84 |
| Mean rel. | 0.104 | 0.119 | 0.088 | 0.107 | 0.078 | 0.098 |
| SD | 0.019 | 0.019 | 0.014 | 0.020 | 0.013 | 0.021 |
| ACo sROI | ||||||
| Mean abs. | 99.90 | 132.82 | 127.64 | 135.08 | 111.80 | 131.20 |
| SD | 35.74 | 27.81 | 42.47 | 31.58 | 34.02 | 23.72 |
| Mean rel. | 0.088 | 0.082 | 0.088 | 0.071 | 0.087 | 0.064 |
| SD | 0.017 | 0.017 | 0.013 | 0.010 | 0.011 | 0.007 |
| Elderly subjects (N=5) | ||||||
Intra-rater reliability and inter-rater reliability for the tracing of the whole amygdala exceeded the ‘almost perfect’ (>0.81) range (Landis and Koch, 1977) for both ICC and Dice coefficients.
The ICC and Dice coefficient measurements for intra-rater and inter-rater reliabilities are presented in Table 2.
Table 2.
Correlation coefficients (ICC) and dice coefficients for left and right hemisphere whole amygdala and subregions in all 10 subjects.
| Intra-rater data (operator JE) |
Inter-rater data |
|||
|---|---|---|---|---|
| L | R | L | R | |
| Whole amygdala | ||||
| Dice coefficient | 0.864 | 0.883 | 0.873 | 0.875 |
| ICC | 0.964 | 0.957 | 0.966 | 0.881 |
| BL sROI | ||||
| Dice coefficient | 0.968 | 0.969 | 0.965 | 0.958 |
| ICC | 0.941 | 0.969 | 0.964 | 0.898 |
| BM sROI | ||||
| Dice coefficient | 0.828 | 0.820 | 0.810 | 0.756 |
| ICC | 0.921 | 0.889 | 0.902 | 0.780 |
| CM sROI | ||||
| Dice coefficient | 0.907 | 0.925 | 0.868 | 0.880 |
| ICC | 0.948 | 0.934 | 0.831 | 0.807 |
| ACo sROI | ||||
| Dice coefficient | 0.895 | 0.868 | 0.851 | 0.835 |
| ICC | 0.948 | 0.893 | 0.937 | 0.652 |
Amygdala subregions
While there were differences between subjects in relatively minor aspects of the size, shape, and orientation of the geometric properties relevant for this protocol, there were no subjects with substantially different anatomic features that would require the use of an alternative variant protocol.
Intra-rater reliability results for the amygdala subregions in both hemispheres were strong. The average Dice coefficient for the subregions was 0.90 for both the left and right hemispheres. For ICC, the average was 0.94 for the left hemisphere and 0.92 for the right hemisphere.
Inter-rater reliability was slightly lower than intra-rater stability. The average Dice coefficient across all subregions was 0.87 for the left hemisphere and 0.86 for the right hemisphere. The average ICC was 0.91 for the left hemisphere and 0.78 for the right hemisphere.
Discussion
Using ultra-high resolution MRI images, we created two reliable segmentation protocols for the human amygdala in vivo. First, we developed a modified approach to identifying whole amygdala boundaries. The high level of detail visible in these images allowed us to easily and reliably discern relevant anatomical landmarks–such as the alveus–that presented more difficulty to past studies using lower-resolution MRIs (Achten et al., 1998; Pruessner et al., 2000; Watson et al., 1992). The level of reliability for this protocol–as defined by the ICC and dice overlap coefficient–is very high (for example, Pruessner et al., 2000 presents inter-rater ICCs of .84 for the left amygdala and .83 for the right amygdala compared to our ratings of 0.97 and 0.88, respectively). Importantly, we were also able to establish a more detailed method for including the dorsal amygdala in its caudal portion–the location of the caudal Ce–than has been documented previously. This reliable visualization of the fine structure of the amygdala facilitated the second experiment, which was the creation of a manual method for segmenting the amygdala into subregions.
We developed a novel method for manually segmenting amygdala subregions approximately corresponding to subnuclei. A geometric approach based on specifically visible landmarks was developed and determined to be reliable. The protocol allows for demarcation of the BL sROI, the BM sROI, the CM sROI, and the ACo sROI. Although it would be ideal to be able to trace fine white matter bands between amygdala subnuclei, these were not reliably visible even in images of the present resolution.
For both of these experiments, we created a detailed visual atlas that presents histological and MRI slices side by side, along with the tracings. We believe that this will have value for future work in which investigators aim to reproduce or refine the present method. Past studies on the whole amygdala have not published such detailed images and therefore it can be difficult to determine the exact implementations of boundaries in previous protocols (Achten et al., 1998; Chupin et al., 2007a, 2007b; Morey et al., 2009a, 2009b). We hope that the level of detail in our second experiment will provide support for the growing interest in the amygdaloid complex as a collection of disparately organized nuclei with distinct anatomical, connectional, and functional properties (Holland and Gallagher, 1999; Swanson and Petrovich, 1998).
The names of the structures labeled in our segmentation protocol generally correspond to those of Amunts et al. (2005), except for one major distinction: we further separated the “basolateral region” into the basolateral subregion (BL SROI) and the basomedial subregion (BM sROI). Traditionally, the nuclei have been analyzed together because of their relatedness in anatomy and connectivity, such as with the visceromotor areas of the insula (e.g. Barbas, 2000) and the subgenual anterior cingulate cortex (sgACC) (Vogt and Miller, 1983; Vogt and Pandya, 1987), as well as to heteromodal regions like the orbitofrontal cortex (OFC) (for a review, see Price, 1999). Nonetheless, recent studies have revealed specialized roles of BL and BM. For example, an anterograde and retrograde tracing study in rats found that the temporal region of hippocampal subiculum/CA1 projects mainly to the basomedial region while the septal portion of subiculum/CA1 sends efferents to the more lateral portions of the amygdala, such as the basolateral nucleus (Kishi et al., 2006). Furthermore, a study of nitric-oxide producing neurons in the basolateral complex of developing mice found that the three regions of the BL complex (the basolateral, lateral, and basomedial) had separate immunoreactive profiles (Olmos et al., 2005). The results of these and other studies provide motivation to attempt to separate BL sROI and BM sROI from each other in a protocol such as the present one.
Another focus of our design was to include a greater portion of the CM, which is particularly unique as the main output hub of the amygdala (Price and Amaral, 1981). It alone has cellular connections to the adjacent bed nucleus of the stria terminalis. The two structures form an anatomical continuum termed the extended amygdala that receives input from but does not include the BL (de Olmos and Heimer, 2006). In addition, the CM has specialized projections to the paraventricular nucleus of the hypothalamus (Marcilhac and Siaud, 1997), dorsal substantia innominata (Bourgeais et al., 2001), the basal nucleus of Meynert, the medial pulvinar nucleus and central nucleus of the thalamus, the parabrachial region of the pons and numerous cortical regions (Price and Amaral, 1981). Reliable measurement of the CM separately from other nuclei is important because of what is thought to be its unique role in attention processes (Holland and Gallagher, 1999; Wheeler and Holland, 2011), social communication (Bamshad et al., 1997), pain processing (Bernard et al., 1992), stress mediation (Yang et al., 2008), motivation (El-Amamy and Holland, 2007), and drug-related behavior (Fu et al., 2007; Rezayof et al., 2007), to name a few.
The fourth amygdala subdivision in our model was the collection of superficial nuclei that include the dorsal, ventral, medial and lateral portions of the ACo. This region has dense connections to the olfactory and accessory olfactory bulb, mPFC, agranular insula, caudal thalamus, perirhinal cortex, caudolateral hypothalamus, and hippocampus (Swanson and Petrovich, 1998).
This study has several limitations. The first limitation is the need to acquire ultra-high resolution data in order to visualize the amygdala and surrounding tissue sufficiently to see the landmarks described; currently, about 35 min of scan time is required to obtain these data. Although this may limit the types of individuals from which data such as this can be acquired, we have managed to obtain these scans in many cognitively normal older individuals as well as individuals with mild dementia. One way we accomplished this is to acquire multiple shorter scans (6–7 min), each of which have relatively low signal-to-noise, and then motion-correct and average them together. Thus, acquisitions corrupted by motion can be discarded. Advances in data acquisition methods, such as parallel imaging (Wiggins et al., 2007), on-line motion correction (van der Kouwe et al., 2005), and higher field strengths (Wiggins et al., 2006), should enable these types of data to be acquired more efficiently. Second, even with this high-resolution data, it is challenging to visualize tissue properties that correspond to subnuclei structure with adjacent white matter bands, which is why we employed a geometric protocol for subregions. Other MR contrasts, such as T2-weighted images or combinations of T1- and T2-weighted images may provide additional information that could be used to visualize these structures more clearly (Solano-Castiella et al., 2011). Given the relatively high content of iron in regions within the amygdala, a T2*-weighted image or possibly the use of an off-resonance magnetization transfer prepulse could be beneficial (Hu et al., 2011; Solano-Castiella et al., 2011; Zivadinov et al., 2012). However, acquiring such data using three-dimensional sequences that are undistorted with respect to each other is challenging, requiring the development of new sequences (e.g., multi-echo MPRAGE) (van der Kouwe et al., 2008). Next, the tracing protocol we have described is time-consuming, taking a trained operator approximately 60 min per subject (both hemispheres). We hope that an automated version of this protocol will be developed to produce more expedient and highly reliable results, as has been done for the hippocampal formation (Van Leemput et al., 2008). In addition, some voxels from structures that are not part of the amygdala are included in our subdivisions (e.g. piriform cortex in the BM SROI subregion). Similarly, some voxels from the ACo are occasionally labeled as BM SROI. These errors are unavoidable in a geometrical protocol but they represent very small fractions of the structural designations. Furthermore, reliability is not fully optimal in some regions, but this limitation is common in published morphometric protocols including small structures (Desikan et al., 2006). Finally, this protocol has not been validated against cytoarchitecturally derived boundaries from post-mortem specimens, but this is a limitation common to most similar studies in the field.
In summary, our novel approaches to defining the boundaries and subregions of the amygdala offer high reliability for measuring these structures in living humans. The protocols may be helpful for studying the anatomical, functional and connectional properties of the amygdala and its subnuclei. Refined analyses of these regions are crucial for understanding normal psychological function and how these functions change with age and pathology.
Supplementary Material
Box 1.
ac, anterior commissure; ACo, amygdaloid cortical nucleus; ACoD, anterior cortical amygdaloid nucleus, dorsal part; ACoV, anterior cortical amygdaloid nucleus, ventral part; AG, ambiens gyrus; Ai, amygdaloid island; alv, alveus; AStr, amygdalostriatal transition area; BC, basal nucleus, compact part; BLD, basolateral amygdaloid nucleus, dorsal (magnocellular) part; BLI, basolateral amygdaloid nucleus, intermediate part; BLPL, basolateral amygdaloid nucleus, paralaminar part; BLVL, basolateral amygdaloid nucleus, ventrolateral part; BLVM, basolateral amygdaloid nucleus, ventromedial part; BM, basomedial amygdaloid nucleus; CA1, CA1 field of hippocampus; Ce, central amygdaloid nucleus; CeL, central amygdaloid nucleus, lateral part; CeM, central amygdaloid nucleus, medial part; CM, centromedial complex; DiCl, diffuse insular claustrum; Ent, entorhinal cortex; HiH, hippocampal head; La, lateral amygdaloid nucleus; LaDA, lateral amygdaloid nucleus, dorsal anterior part; LaDL, lateral amygdaloid nucleus, dorsolateral part; LaDM, lateral amygdaloid nucleus, dorsomedial part; Lal, lateral amygdaloid nucleus, intermediate part; LaV, lateral amygdaloid nucleus, anterior part; LiCl, limitans claustrum; Me, medial amygdaloid nucleus; MeA, medial amygdaloid nucleus, ant. part; opt, optic tract; PaCl, preamygdalar claustrum; PAM, periamygdaloid cortex; PCo, post. cortical amygdaloid nucleus; PHA, parahippocampal–amygdaloid transition area; PHG, parahippocampal gyrus; PirT, piriform cortex, temporal area; Pu, putamen; PuV, ventral putamen; S, subiculum; sas, semiannular sulcus; SEpS, subependymal stratum; SLG, semilunar gyrus; st, stria terminalis; TCl, temporal claustrum; TLV, temporal horn of lateral ventricle; Un, uncus; unc, uncinate fasciculus; us, uncal sulcus; VCl, ventral claustrum.
Acknowledgments
This work was supported by grants from the National Institute on Aging (R01 AG029840) and the Alzheimer’s Association to Dr. Brad Dickerson and from the National Institutes of Health Director’s Pioneer Award (DP1OD003312) and a National Institute on Aging grant (AG030311) to Lisa Feldman Barrett. The authors thank Michael Brickhouse for excellent technical assistance.
Footnotes
Appendix A.Supplementary data Supplementary data to this article can be found online at doi:10.1016/j.neuroimage.2011.12.073.
References
- Achten E, Deblaere K, De Wagter C, Van Damme F, Boon P, De Reuck J, Kunnen M. Intra- and interobserver variability of MRI-based volume measurements of the hippocampus and amygdala using the manual ray-tracing method. Neuroradiology. 1998;40(9):558–566. doi: 10.1007/s002340050644. [DOI] [PubMed] [Google Scholar]
- Adolphs R, Tranel D, Damasio H, Damasio A. Impaired recognition of emotion in facial expressions following bilateral damage to the human amygdala. Nature. 1994;372(6507):669–672. doi: 10.1038/372669a0. [DOI] [PubMed] [Google Scholar]
- Amaral DG. The amygdala, social behavior, and danger detection. Ann. N. Y. Acad. Sci. 2003;1000:337–347. doi: 10.1196/annals.1280.015. [DOI] [PubMed] [Google Scholar]
- Amunts K, Kedo O, Kindler M, Pieperhoff P, Mohlberg H, Shah NJ, Habel U, Schneider F, Zilles K. Cytoarchitectonic mapping of the human amygdala, hippocampal region and entorhinal cortex: intersubject variability and probability maps. Anat. Embryol. (Berl.) 2005;210(5–6):343–352. doi: 10.1007/s00429-005-0025-5. [DOI] [PubMed] [Google Scholar]
- Anand A, Shekhar A. Brain imaging studies in mood and anxiety disorders: special emphasis on the amygdala. Ann. N. Y. Acad. Sci. 2003;985:370–388. doi: 10.1111/j.1749-6632.2003.tb07095.x. [DOI] [PubMed] [Google Scholar]
- Bach DR, Behrens TE, Garrido L, Weiskopf N, Dolan RJ. Deep and superficial amygdala nuclei projections revealed in vivo by probabilistic tractography. J. Neurosci. 2011;31(2):618–623. doi: 10.1523/JNEUROSCI.2744-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad M, Karom M, Pallier P, Albers HE. Role of the central amygdala in social communication in Syrian hamsters (Mesocricetus auratus) Brain Res. 1997;744(1):15–22. doi: 10.1016/s0006-8993(96)01061-x. [DOI] [PubMed] [Google Scholar]
- Barbas H. Connections underlying the synthesis of cognition, memory, and emotion in primate prefrontal cortices. Brain Res. Bull. 2000;52(5):319–330. doi: 10.1016/s0361-9230(99)00245-2. [DOI] [PubMed] [Google Scholar]
- Barrett LF, Bliss-Moreau E, Duncan SL, Rauch SL, Wright CI. The amygdala and the experience of affect. Soc. Cogn. Affect. Neurosci. 2007;2(2):73–83. doi: 10.1093/scan/nsl042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard JF, Huang GF, Besson JM. Nucleus centralis of the amygdala and the globus pallidus ventralis: electrophysiological evidence for an involvement in pain processes. J. Neurophysiol. 1992;68:551–569. doi: 10.1152/jn.1992.68.2.551. [DOI] [PubMed] [Google Scholar]
- Bonilha L, Kobayashi E, Cendes F, Min Li L. Protocol for volumetric segmentation of medial temporal structures using high-resolution 3-D magnetic resonance imaging. Hum. Brain Mapp. 2004;22(2):145–154. doi: 10.1002/hbm.20023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeais L, Gauriau C, Bernad J. Projections from the nociceptive area of the central nucleus of the amygdala to the forebrain: a PHA-L study in the rat. Eur. J. Neurosci. 2001;14(2):229–255. doi: 10.1046/j.0953-816x.2001.01640.x. [DOI] [PubMed] [Google Scholar]
- Chupin M, Hammers A, Bardinet E, Colliot O, Liu RS, Duncan JS, Garnero L, Lemieux L. Fully automatic segmentation of the hippocampus and the amygdala from MRI using hybrid prior knowledge. Med. Image Comput. Comput. Assist. Interv. 2007a;10(1):875–882. doi: 10.1007/978-3-540-75757-3_106. [DOI] [PubMed] [Google Scholar]
- Chupin M, Mukuna-Bantumbakulu AR, Hasboun D, Bardinet E, Baillet Kinkingnéhun S, Lemieux L, Dubois B, Garnero L. Anatomically constrained region deformation for the automated segmentation of the hippocampus and the amygdala: method and validation on controls and patients with Alzheimer’s disease. NeuroImage. 2007b;34(3):996–1019. doi: 10.1016/j.neuroimage.2006.10.035. [DOI] [PubMed] [Google Scholar]
- Convit A, McHugh P, Wolf OT, de Leon MJ, Bobinski M, De Santi S, Roche A, Tsui W. MRI volume of the amygdala: a reliable method allowing separation from the hippocampal formation. Psychiatry Res. 1999;90(2):113–123. doi: 10.1016/s0925-4927(99)00007-4. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I: segmentation and surface reconstruction. Neuroimage. 1999;9(2):179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Davis M, Walker DL, Miles L, Grillon C. Phasic vs sustained fear in rats and humans: role of the extended amygdala in fear vs anxiety. Neuropsychopharmacology. 2010;35(1):105–135. doi: 10.1038/npp.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Olmos JS, Heimer L. The concepts of the ventral striatopallidal system and extended amygdala. Ann. N. Y. Acad. Sci. 2006;877:1–32. doi: 10.1111/j.1749-6632.1999.tb09258.x. [DOI] [PubMed] [Google Scholar]
- Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302. [Google Scholar]
- El-Amamy H, Holland PC. Dissociable effects of disconnecting amygdala central nucleus from the ventral tegmental area or substantia nigra or learned orienting and incentive motivation. Eur. J. Neurosci. 2007;25(5):1557–1567. doi: 10.1111/j.1460-9568.2007.05402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadok JP, Darvas M, Dickerson TM, Palmiter RD. Long-term memory for Pavlovian fear conditioning requires dopamine in the nucleus accumbens and basolateral amygdala. PLoS One. 2010;5(9):e12751. doi: 10.1371/journal.pone.0012751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc. Natl. Acad. Sci. USA. 2000;97(20):11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Tootell RB, Dale AM. High-resolution intersubject averaging and a coordinate system for the cortical surface. Hum. Brain Mapp. 1999a;8(4):272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999b;9(2):195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fischl B, Liu A, Dale AM. Automated manifold surgery: constructing geomet rically accurate and topologically correct models of the human cerebral cortex. IEEE Trans. Med. Imaging. 2001;20(1):70–80. doi: 10.1109/42.906426. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale M. Whole brain segmentation: auto\mated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Fu Y, Pollandt S, Lui J, Krishnan B, Genzer K, Orozco-Cabal L, Gallagher JP, Shinnick-Gallagher P. Long-term potentiation (LTP) in the central amygdala (CeA) is enhanced after prolonged withdrawal from chronic cocaine and requires CRF1 receptors. J. Neurophysiol. 2007;97(1):937–941. doi: 10.1152/jn.00349.2006. [DOI] [PubMed] [Google Scholar]
- Gaffan D, Gaffan EA, Harrison S. Visual–visual associative learning and reward-association learning in monkeys: the role of the amygdala. J. Neurosci. 1989;9(2):558–564. doi: 10.1523/JNEUROSCI.09-02-00558.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland PC, Gallagher M. Amygdala circuitry in attentional and representational processes. Trends Cogn. Sci. 1999;3(2):65–73. doi: 10.1016/s1364-6613(98)01271-6. [DOI] [PubMed] [Google Scholar]
- Hooker CI, Germine LT, Knight RT, D’Esposito M. Amygdala response to facial expressions reflects emotional learning. J. Neurosci. 2006;26(35):8915–8922. doi: 10.1523/JNEUROSCI.3048-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Coupe P, Pruessner JC, Collins DL. Appearance-based modeling for segmentation of hippocampus and amygdala using multi-contrast MR imaging. NeuroImage. 2011;58:549–559. doi: 10.1016/j.neuroimage.2011.06.054. [DOI] [PubMed] [Google Scholar]
- Kishi T, Tsumori T, Yokota S, Yasui Y. Topographical projection from the hippocampal formation to the amygdala: a combined anterograde and retrograde tracing study in the rat. J. Comp. Neurol. 2006;496(3):349–368. doi: 10.1002/cne.20919. [DOI] [PubMed] [Google Scholar]
- Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159–174. [PubMed] [Google Scholar]
- Lanteaume L, Khalfa S, Régis J, Marquis P, Chauvel P, Bartolmei F. Emotion induction after direct intracerebral stimulations of human amygdala. Cereb. Cortex. 2007;17(6):1307–1313. doi: 10.1093/cercor/bhl041. [DOI] [PubMed] [Google Scholar]
- LeDoux JE, Iwata J, Cicchetti P, Reis DJ. Different projections of the central amygdaloid nucleus mediate autonomic and behavioral correlates of conditioned fear. J. Neurosci. 1988;8(7):2517–2529. doi: 10.1523/JNEUROSCI.08-07-02517.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado CJ, Emery NJ, Capitanio JP, Mason WA, Mendoza SP, Amaral DG. Bilateral neurotoxic amygdala lesions in rhesus monkeys (Macaca mulatta): consistent pattern of behavior across different social contexts. Behav. Neurosci. 2008;22(2):251–266. doi: 10.1037/0735-7044.122.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai J, Paxinos G, Voss T. Atlas of the Human Brain. 3rd ed Elsevier Academic Press; London: 1997. [Google Scholar]
- Makris N, Meyer JW, Bates JF, Yeterian EH, Kennedy DN, Caviness VS. MRI-based topographic parcellation of human cerebral white matter and nuclei II. Rationale and applications with systematics of cerebral connectivity. NeuroImage. 1999;9(1):18–45. doi: 10.1006/nimg.1998.0384. [DOI] [PubMed] [Google Scholar]
- Marcilhac A, Siaud P. Identification of projections from the central nucleus of the amygdala to the paraventricular nucleus of the hypothalamus which are immunoreactive for corticotrophin-releasing hormone in the rat. Exp. Physiol. 1997;82(2):273–281. doi: 10.1113/expphysiol.1997.sp004022. [DOI] [PubMed] [Google Scholar]
- Matsuoka Y, Mori E, Inagaki M, Kozaki Y, Nakano T, Wenner M, Uchitomi Y. Manual tracing guideline for volumetry of hippocampus and amygdala with high-resolution MRI. No To Shinkei. 2003;55(8):690–697. [PubMed] [Google Scholar]
- Morey RA, Petty CM, Xu Y, Hayes JP, Wagner HR, II, Lewis DV, LaBar KS, Styner M, McCarthy G. A comparison of automated segmentation and manual tracing for quantifying hippocampal and amygdala volumes. NeuroImage. 2009a;45(3):855–866. doi: 10.1016/j.neuroimage.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey RA, Petty CM, Xu Y, Hayes JP, Wagner HR, II, Lewis DV, Labar KS, Styner M, McCarthy G. Rebuttal to Hasan and Pedraza in comments and controversies: “improving the reliability of manual and automated methods for hippocampal and amygdala volume measurements”. NeuroImage. 2009b;48(3):499–500. doi: 10.1016/j.neuroimage.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JS, Ohman A, Dolan RJ. Conscious and unconscious emotional learning in the human amygdala. Nature. 1998;393(6684):467–470. doi: 10.1038/30976. [DOI] [PubMed] [Google Scholar]
- Olmos JL, Real MA, Medina L, Guirado S, Dávila JC. Distribution of nitric oxide-producing neurons in the developing and adult mouse amygdalar basolateral complex. Brain Res. Bull. 2005;66(4–6):465–469. doi: 10.1016/j.brainresbull.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Packard MG, Cahill L, McGaugh JL. Amygdala modulation of hippocampal-dependent and caudate nucleus-dependent memory processes. Proc. Natl. Acad. Sci. U. S. A. 1994;91(18):8477–8481. doi: 10.1073/pnas.91.18.8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Mai J. The Human Nervous System. 2nd ed Elsevier Academic Press; San Diego: 2004. [Google Scholar]
- Pessoa L, Kastner S, Ungerleider LG. Attentional control of the processing of neural and emotional stimuli. Brain Res. Cogn. Brain Res. 2002a;15(1):31–45. doi: 10.1016/s0926-6410(02)00214-8. [DOI] [PubMed] [Google Scholar]
- Pessoa L, McKenna M, Gutierrez E, Ungerleider LG. Neural processing of emotional faces requires attention. Proc. Natl. Acad. Sci. U. S. A. 2002b;99(17):11458–11463. doi: 10.1073/pnas.172403899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps EA, LeDoux JE. Contributions of the amygdala to emotion processing: from animal models to human behavior. Neuron. 2005;48:175–187. doi: 10.1016/j.neuron.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Price JL. Prefrontal cortical networks related to visceral function and mood. Ann. N. Y. Acad. Sci. 1999;877:383–396. doi: 10.1111/j.1749-6632.1999.tb09278.x. [DOI] [PubMed] [Google Scholar]
- Price JL, Amaral DG. An autoradiographical study of the projections of the central nucleus of the monkey amygdala. J. Neurosci. 1981;1(11):1242–1259. doi: 10.1523/JNEUROSCI.01-11-01242.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruessner JC, Li LM, Serles W, Pruessner M, Collins DL, Kabani N, Lupien S, Evans AC. Volumetry of hippocampus and amygdala with high-resolution MRI and three-dimensional analysis software: minimizing the discrepancies between laboratories. Cereb. Cortex. 2000;10(4):433–442. doi: 10.1093/cercor/10.4.433. [DOI] [PubMed] [Google Scholar]
- Rezayof A, Golhasani-Keshtan F, Haeri-Rohani A, Zarringdast MR. Morphine-induced place preference: involvement of the central amygdala NMDA receptors. Brain Res. 2007;1133(1):34–41. doi: 10.1016/j.brainres.2006.11.049. [DOI] [PubMed] [Google Scholar]
- Rosvold HE, Mirsky AF, Pribram KH. Influence of amygdalectomy on social behavior in monkeys. J. Comp. Physiol. Psychol. 1954;47(3):173–178. doi: 10.1037/h0058870. [DOI] [PubMed] [Google Scholar]
- Roy AK, Shehzad Z, Margulies DS, Kelly AM, Uddin LQ, Gotimer K, Biswal BB, Castellanos FX, Milham MP. Functional connectivity of the human amygdala using resting state fMRI. NeuroImage. 2009;45(2):614–626. doi: 10.1016/j.neuroimage.2008.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander K, Scheich H. Auditory perception of laughing and crying activates human amygdala regardless of attentional state. Brain Res. Cogn. Brain Res. 2001;12(2):181–198. doi: 10.1016/s0926-6410(01)00045-3. [DOI] [PubMed] [Google Scholar]
- Saygin ZM, Osher DE, Augustinack J, Fischl B, Gabrieli JD. Connectivity-based segmentation of human amygdala nuclei using probabilistic tractography. Neuroimage. 2011;56(3):1353–1361. doi: 10.1016/j.neuroimage.2011.03.006. Epub ahead of print 2011 Mar 8, from http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WNP-52BGCSF-3&_user=10&_coverDate=06%2F01%2F2011&_rdoc=1&_fmt=high&_orig=gateway&_origin=gateway&_sort=d&_docanchor=&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=950c98628d74c13c4304dae864458390&searchtype=a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ségonne F, Dale AM, Busa E, Glessner M, Salat D, Hahn HK, Fischl B. A hybrid approach to the skull stripping problem in MRI. Neuroimage. 2004;22(3):1060–1075. doi: 10.1016/j.neuroimage.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Sharot T, Riccardi AM, Raio CM, Phelps EA. Neural mechanisms mediating optimism bias. Nature. 2007;450(7166):102–105. doi: 10.1038/nature06280. [DOI] [PubMed] [Google Scholar]
- Shrout PE, Fleiss JL. Intraclass correlations: Uses in assessing rater reliability. Psychol. Bull. 1979;86(2):420–428. doi: 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- Solano-Castiella E, Anwander A, Lohmann G, Weiss M, Docherty C, Geyer S, Reimer E, Friederici AD, Turner R. Diffusion tensor imaging segments the human amygdala in-vivo. NeuroImage. 2010;49(2010):2958–2965. doi: 10.1016/j.neuroimage.2009.11.027. [DOI] [PubMed] [Google Scholar]
- Solano-Castiella E, Schafer A, Reimer E, Turke E, Proger T, Lohmann G, Trampel R, Turner R. Parcellation of human amygdala in vivo using ultra high field structural MRI. NeuroImage. 2011;58:741–748. doi: 10.1016/j.neuroimage.2011.06.047. [DOI] [PubMed] [Google Scholar]
- Swanson LW, Petrovich GD. What is the amygdala? Trends Neurosci. 1998;21(8):323–331. doi: 10.1016/s0166-2236(98)01265-x. [DOI] [PubMed] [Google Scholar]
- Ursin H, Kaada BR. Subcortical structures mediating the attention response induced by amygdala stimulation. Exp. Neurol. 1960;2:109–122. doi: 10.1016/0014-4886(60)90002-9. [DOI] [PubMed] [Google Scholar]
- van der Kouwe AJ, Benner T, Fischl B, Schmitt F, Salat DH, Harder M, Sorensen AG, Dale AM. On-line automatic slice positioning for brain MR imaging. NeuroImage. 2005;27:222–230. doi: 10.1016/j.neuroimage.2005.03.035. [DOI] [PubMed] [Google Scholar]
- van der Kouwe AJ, Benner T, Salat DH, Fischl B. Brain morphometry with multiecho MPRAGE. NeuroImage. 2008;40:559–569. doi: 10.1016/j.neuroimage.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leemput K, Bakkour A, Benner T, Wiggins G, Wald LL, Augustinack J, Dickerson BC, Golland P, Fischl B. Model-based segmentation of hippocampal subfields in ultra-high resolution in vivo MRI. Med. Image Comput. Comput. Assist. Interv. 2008;11:235–243. doi: 10.1007/978-3-540-85988-8_29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leemput K, Bakkour A, Benner T, Wiggins G, Wald LL, Augustinack J, Dickerson BC, Golland P, Fischl B. Automated segmentation of hippocampal subfields from ultra-high resolution in vivo MRI. Hippocampus. 2009;19(6):549–557. doi: 10.1002/hipo.20615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt BA, Miller MW. Cortical connections between rat cingulate cortex and visual, motor and postsubicular cortices. J. Comp. Neurol. 1983;216(2):192–210. doi: 10.1002/cne.902160207. [DOI] [PubMed] [Google Scholar]
- Vogt BA, Pandya DN. Cingulate cortex of the rhesus monkey: II. Cortical afferents. J. Comp. Neurol. 1987;262(2):271–289. doi: 10.1002/cne.902620208. [DOI] [PubMed] [Google Scholar]
- Watson C, Andermann F, Gloor P, Jones-Gotman M, Peters T, Evans A, Olivier A, Melanson D, Leroux G. Anatomic basis of amygdaloid and hippocampal volume measurement by magnetic resonance imaging. Neurology. 1992;42(9):1743–1750. doi: 10.1212/wnl.42.9.1743. [DOI] [PubMed] [Google Scholar]
- Whalen PJ, Rauch SL, Etcoff NL, McInerney SC, Lee MB, Jenike MA. Masked presentations of emotional facial expressions modulate amygdala activity without explicit knowledge. J. Neurosci. 1998;18(1):411–418. doi: 10.1523/JNEUROSCI.18-01-00411.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen PJ, Shin LM, McInerney SC, Fischer H, Wright CI, Rauch SL. A functional MRI study of human amygdala responses to facial expressions of fear versus anger. Emotion. 2001;1(1):70–83. doi: 10.1037/1528-3542.1.1.70. [DOI] [PubMed] [Google Scholar]
- Wheeler DS, Holland PC. Effects of reward timing information on cue associability are mediated by amygdala central nucleus. Behav. Neurosci. 2011;125(1):46–53. doi: 10.1037/a0021951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggins GC, Kraff O, Zakszewski E, Alagappan V, Wiggins CJ, Wald LL. A 7 Tesla gradient mode birdcage coil for improved temporal and occipital lobe SNR. International Society for Magnetic Resonance in Medicine. 2006 [Google Scholar]
- Wiggins GC, Alagappan V, Potthast A, Schmitt M, Wiggins CJ, Fischer H, Jahns K, Benner T, Polimeni JR, Wald LL. Design optimization and SNR performance of 3T 96 channel phased array head coils. Magn. Reson. Med. 2007;62(3):754–762. doi: 10.1002/mrm.22028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CI. The human amygdala in normal aging and Alzheimer’s disease. In: Whalen PJ, Phelps EA, editors. The Human Amygdala. The Guilford Press; New York: 2009. pp. 382–405. [Google Scholar]
- Yang CH, Huang CC, Hsu KS. Differential roles of the basolateral and central amygdala on the effects of uncontrollable stress on hippocampal synaptic plasticity. Hippocampus. 2008;18(6):548–563. doi: 10.1002/hipo.20414. [DOI] [PubMed] [Google Scholar]
- Yassa MA, Stark CEL. A quantitative evaluation of cross-participant registration techniques for MRI studies of the medial temporal lobe. NeuroImage. 2009;44(2):319–327. doi: 10.1016/j.neuroimage.2008.09.016. [DOI] [PubMed] [Google Scholar]
- Zivadinov R, Heininen-Brown M, Schirda CV, Poloni GU, Bergsland N, Magnano CR, Durfee J, Kennedy C, Carl E, Hagemeier J, Benedict RH, Weinstock-Guttman B, Dwyer MG. Abnormal subcortical deep-gray matter susceptibility-weighted imaging filtered phase measurements in patients with multiple sclerosis: a case–control study. Neuroimage. 2012;59:331–339. doi: 10.1016/j.neuroimage.2011.07.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.