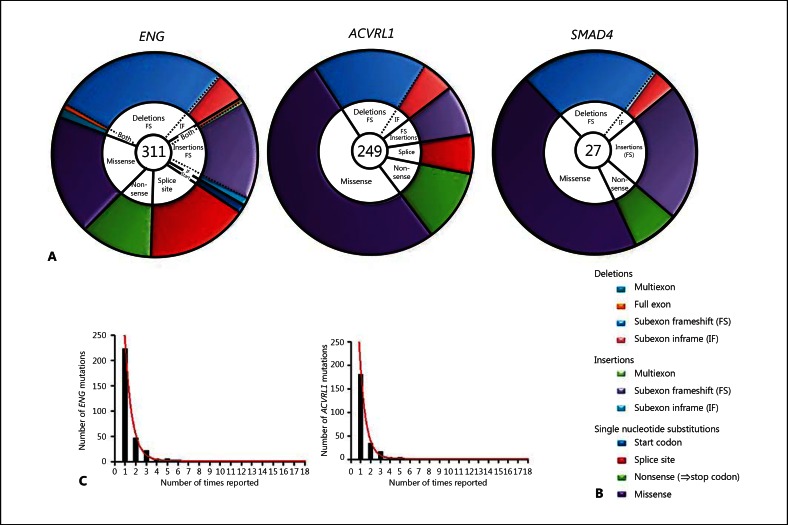
Fig. 1.
Mutational types in HHT. A, B Comparison for all known HHT genes with pie chart representation of the different mutational types. The number in the centre of each pie refers to the total number of mutations reported at the time of analysis. The colour-coding explained in the schematic key in B is used to illustrate the relative prevalence of the different mutational types and to emphasise different genomic DNA mutational types with similar transcript consequences: single nucleotide substitutions are sub-classified conventionally as missense, nonsense, splice site and start codon mutations; deletions and insertions are sub-classified to emphasise the proportion generating nonsense codons and include complex rearrangements. Nonsense codons are generated by all direct nonsense single nucleotide substitutions (green); all subexon frameshift (FS) deletions (pale blue) and insertions (lilac) but no subexon inframe (IF) insertions or deletions; approximately two-thirds of multiexon and full exon deletions, insertions and rearrangements (aqua, orange and pale green), and approximately two-thirds of splice site mutations (red). The formal assignment of splice site mutations and multiexon deletions to frameshift or non-frameshift groupings depends upon evidence of the mutant endoglin RNA species generated – activation of cryptic splice sites may generate an inframe, detectable RNA species [Shovlin et al., 1997] instead of a more obvious exon skip that would lead to a frameshift. C The distribution of the number of times each individual ENG or ACVRL1 mutation was reported (illustrated as columns), with the respective best fit one phase exponential decay model illustrated.