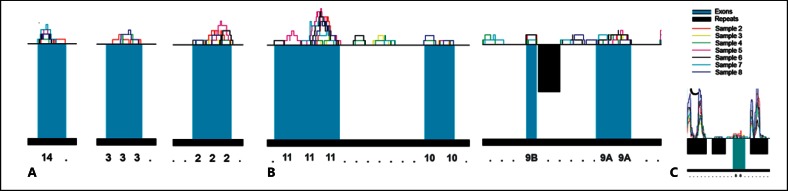
Fig. 5.
Transcribed fragments across regions of the ENG locus deleted in HHT mutations. Colour-coded R-plot data for 7 endothelial cell libraries, with exons represented by blue boxes, repeat regions by black boxes, and individual library alignments by the coloured lines (key, samples 2-8). Note ENG is on the minus strand, so 5′ to 3′ is represented right to left. A Replicate alignments to exons 2, 3 and 14. B Alignments to regions spanning exon 9A-11, demonstrating replicate alignments to exons and to non-repetitive intronic regions. C Illustration of non-unique alignments to repetitive regions (including the Alu elements responsible for a 2-kb ENG deletion spanning exon 8 [Shovlin et al., 1997]). Note that the transcripts responsible for these very high alignments to repetitive regions will have originated throughout the genome and are not necessarily from the ENG locus.