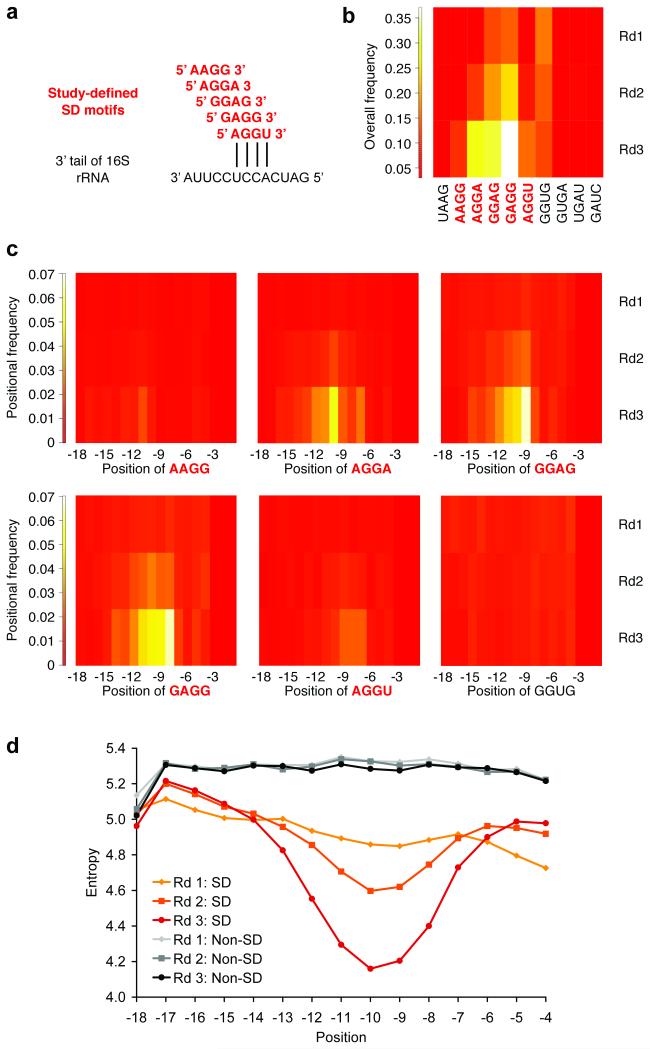
Figure 2.
Enrichment of SD sequences. a) The alignment of study-defined SD motifs (red) with the 3′ tail of the 16S rRNA (black) is shown. b) Overall enrichment of SD sequences over three rounds (Rd1, Rd2, and Rd3) is shown. For comparison, we present all 10 four-base subsets of the reverse complement (5′-UAAGGAGGUGAUC-3′) to the 13 unpaired bases at the 3′ end of the 16S rRNA (5′-GAUCACCUCCUUA-3′) in our selected sequences: UAAG, AAGG, AGGA, GGAG, GAGG, AGGU, GGUG, GUGA, UGAU, and GAUC. c) All study-defined SD motifs (AAGG, AGGA, GGAG, GAGG, and AGGU) exhibited position-dependent enrichment according to their alignment with the 16S rRNA. GGUG (also shown) was enriched overall, but not in a position-dependent manner. d) The position-dependent entropy of four-base windows of 18-base sequences with or without an SD motif over three rounds is shown. Similar plots were observed for five-, six-, seven-, and eight-base windows. Position is specified by the first base of the motif relative to the start codon. SD, Shine-Dalgarno.