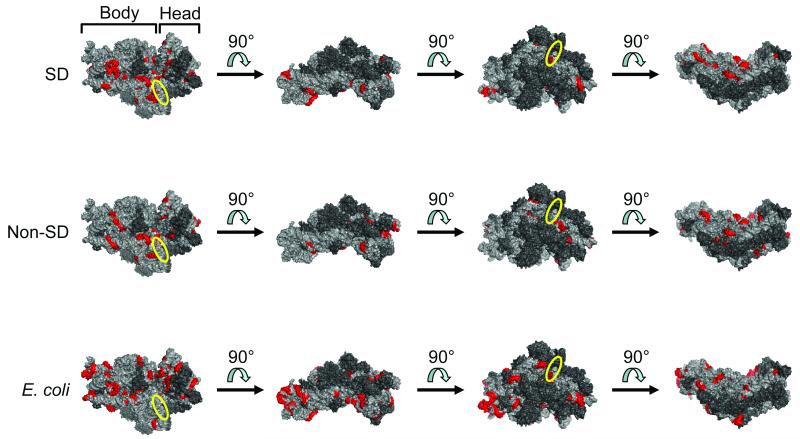
Figure 4.
Distribution of potential sites for base-pairing of RBSs (selected library [SD only], top row; selected library [non-SD only], middle row; and E. coli, bottom row) to 16S rRNA. Regions on the E. coli 30S ribosomal subunit with significant complementarity to the RBS population of interest (p-value < 0.01; Bonferroni-corrected) were determined. Significant six-base windows that shared five bases with at least one neighboring significant window are highlighted in red (PyMOL rendering of PDB 3DF1). Four different views convey the general distribution of these potential base-pairing sites over the small ribosomal subunit. The first view in each row shows the face that becomes buried after assembly with the large ribosomal subunit. The approximate position of the anti-SD sequence is indicated by the yellow ellipse. 16S rRNA = light gray; ribosomal proteins = dark gray. RBS, ribosome binding site; SD, Shine-Dalgarno.