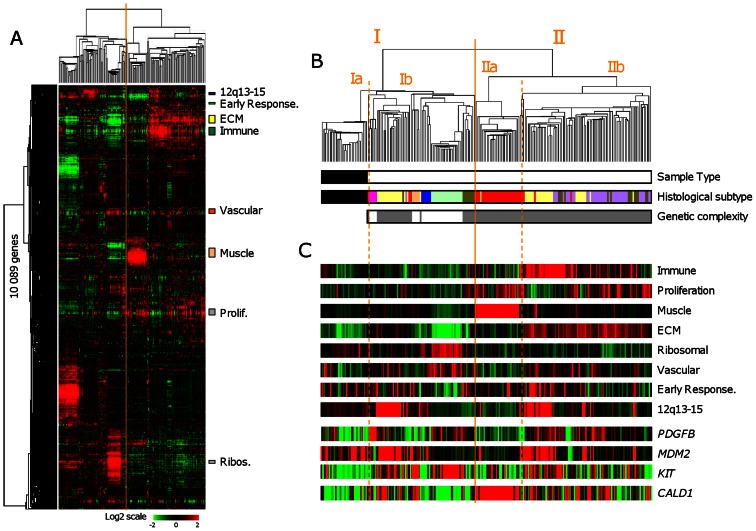
Figure 1. Whole-genome expression profiling of SFTs and STSs.
A. Hierarchical clustering of 208 samples (29 SFTs and 179 STSs) and 10,089 genes with significant variation in mRNA expression level across the samples (SD≥0.25). Each row of the data matrix represents a gene and each column a sample. Expression levels are depicted according to the color scale shown at the bottom, where red and green indicate expression levels respectively above and below the median and the color saturation represents the magnitude of deviation from the median. The dendrogram of samples (above matrixes) represents overall similarities in gene expression profiles and is zoomed in B. Colored bars to the right indicate the locations of 8 gene clusters of interest. B. Dendrogram of samples. Top, two large groups of samples (designated I and II) are evidenced by clustering and delimited by the orange vertical line. Each cluster is divided into two subclusters (a and b) delimited by orange dotted vertical lines. Bottom, some characteristics of samples are represented according to a color ladder: sample type (black, SFT; white, STS); histological subtype (black, SFT; pink, DFSP; salmon, GIST; dark blue, synovial sarcoma; light green, myxoid/round cells liposarcoma; yellow, dedifferentiated liposarcoma; dark green, pleomorphic liposarcoma; red, leiomyosarcoma; purple, malignant fibrous histiocytoma); degree of genetic complexity of STSs (white, simple; grey, complex). C. Metagene (MG) of the 8 gene clusters shown in Figure 1A (the metagene is the mean expression level of included genes) and four control genes (EntrezGene symbol) known as differentially expressed according to the histological subtype.