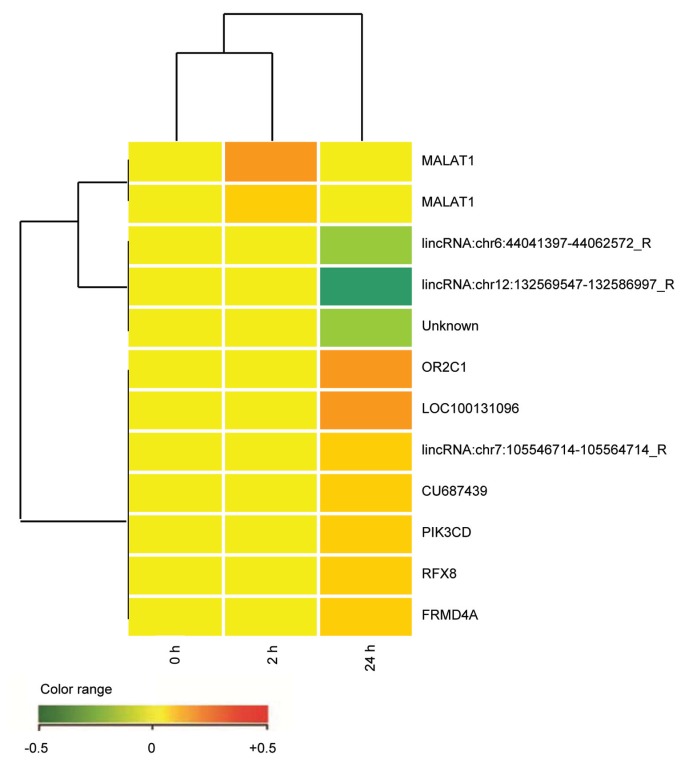
Figure 2.
Comparative microarray analysis of the DL06 cells after CD38 ligation (2 h versus 24 h). RNA was obtained from the DL06 cells incubated at 37°C with anti-CD38 (clone IB4) mAb for 2 h and 24 h. An irrelevant isotype-matched anti-TCRvβ3 mAb was used as control for the same interval times. Four independent experimental settings were analyzed using the two-color comparative hybridization procedure in three cases and the one color protocol in one. mRNA amplification and labeling was followed by hybridization on 8×60K Human Whole Genome Oligo Microarrays (Agilent Technologies, Santa Clara, CA, USA) with technical replication (details in [68]). Images were analyzed using Feature Extraction software v10.5 (Agilent Technologies) and raw data processed within R statistical environment, using the limma library (http://www.bioconductor.org/packages/2.11/bioc/html/limma.html). The raw intensity values were background-corrected with the normexp method and normalized with the quantile method, in the case of one-color hybridization. Two-color hybridization arrays were first subjected to Loess intraarray normalization and then to quantile interarray normalization. For each treatment time, technical replicates were combined and the empirical Bayes method was applied to retrieve any modulated probe in treated versus control comparisons (Benjamini-Hochberg corrected p value <0.01). Each point refers to 1 of the 14 transcripts with adjusted p value ≤0.01 in at least one of the experimental samples analyzed. Red represents upregulated expression and green downregulated expression.