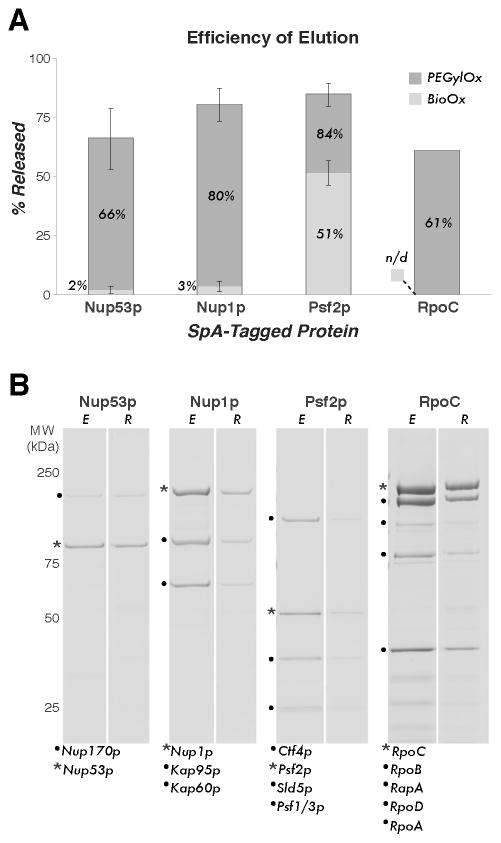
Figure 2. Efficiency of elution of endogenous protein complexes by competitive displacement using Bio-Ox or PEGylOx.
(A) The percentage of S. aureus Protein A (SpA)-tagged protein released from immobilized rabbit IgG after treatment with Bio-Ox or PEGylOx for 15 min at room temperature (RT). At least three experimental replicates were averaged where error bars are shown, indicating the standard deviation (SD). The value for RpoC-SpA release was determined from a single experiment using PEGylOx only (n/d: Bio-Ox not done). All values determined by image densitometry of Coomassie blue stained SDS-PAGE gels (Supplementary Section S3). (B) Example Coomassie blue stained gels showing elution of protein complexes using PEGylOx for each of the proteins quantified in (A). The SpA tagged protein is marked with an asterisk (*), the eluted fractions are denoted E and the retained fractions are denoted R. Displayed are the S. cerevisiae Nup53p/Nup170p dimer (13), Nup1p/Kap95p/Kap60p trimer (13), and pentameric GINS complex (14), and the E. coli RNA polymerase holoenzyme (15). All complexes were prepared from ~0.5 g of cell material. Co-precipitating proteins are labeled based on the referenced work and MALDI-MS analyses (data not shown).