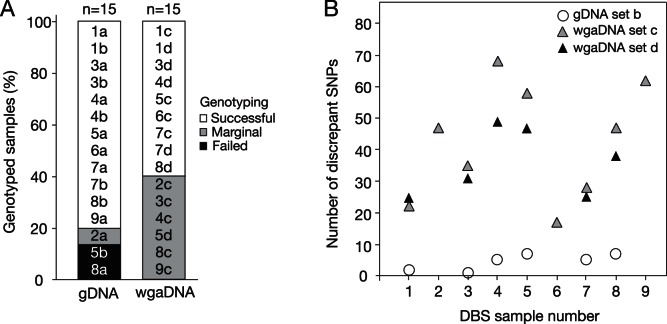
Figure 1. Success and accuracy of genotyping with genomic or amplified DNA from dried blood spots.
Genomic DNA (gDNA) extracted from two 2 mm punches obtained from archived dried blood spots (DBS) from nine individuals (1–9) was genotyped on an Illumina Infinium 300,000 SNP test chip (set “a”, 1a–9a). For six of the DBS (1,3,4,5,7,8), a set (set “b”) of extraction duplicates (duplicate extractions from the same DBS) was prepared and genotyped in the same way. A 1 µl aliquot of each of these 15 samples of gDNA was whole genome amplified (wgaDNA) and then genotyped in the same way as sets “a” and “b”: set “c” wgaDNA samples (1c–9c) were amplified from set “a” gDNA samples, and set “d” wgaDNA samples (1d, 3d, 4d, 5d, 7d, 8d) were amplified from set “b” gDNA samples. (A) Performance quality of the gDNA and wgaDNA samples on genome-wide SNP genotyping. Successful, Marginal, and Failed sample performance parameters are given in Results. (B) SNP genotyping replication error. The number of SNP discrepancies for each sample was determined using gDNA set “a” as the reference. Replication error (the number of discrepancies among SNPs called in both the sample and reference divided by the total number of SNPs called in both) was 1.6×10−5 for set “b”, 1.5×10−4 for set “c”, and 1.2×10−4 for set “d”. These replication error values are provisional because we do not know the true genotype of the discrepant loci and there may be gDNA-specific and/or wgaDNA-specific artifacts.