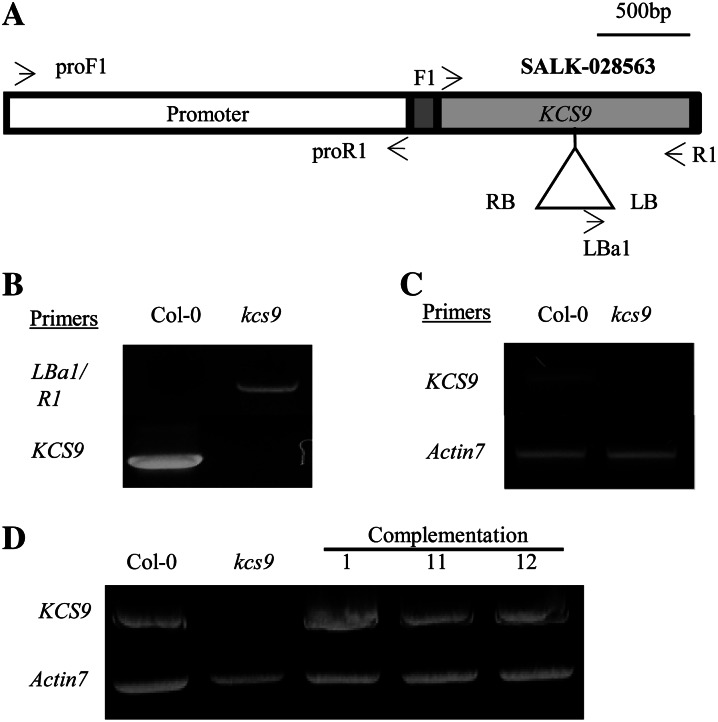
Figure 3.
Isolation of T-DNA-inserted kcs9 mutant (A–C) and generation of complementation lines of the kcs9 mutant (D). A, Genomic organization of the T-DNA-tagged KCS9 gene. T-DNA-inserted kcs9 seeds were obtained from SALK (SALK 028563). The promoter region, 5′ and 3′ untranslated regions, and coding region of the KCS9 gene are shown in the white box, gray boxes, and black box, respectively. LB, Left border; RB, right border. B, Genomic DNA was isolated from the wild type (Col-0) and the kcs9 mutant, and T-DNA insertion was confirmed by genomic DNA PCR using the LBa1 and R1 primers shown in Supplemental Table S1. C, The levels of KCS9 transcripts in 10-d-old seedlings of the wild type (Col-0) and the kcs9 mutant were analyzed by RT-PCR using the F1 and R1 primers shown in Supplemental Table S1. Actin7 was used to determine the quantity and quality of the cDNAs. D, The binary vector harboring KCS9::eYFP under the control of the CaMV 35S promoter was transformed into Arabidopsis for complementation of the kcs9 mutant. The transgenic seedlings were selected on 1/2 MS medium with kanamycin, and leaves of 3-week-old plants were used for RNA isolation to analyze the expression of KCS9 by RT-PCR analysis.