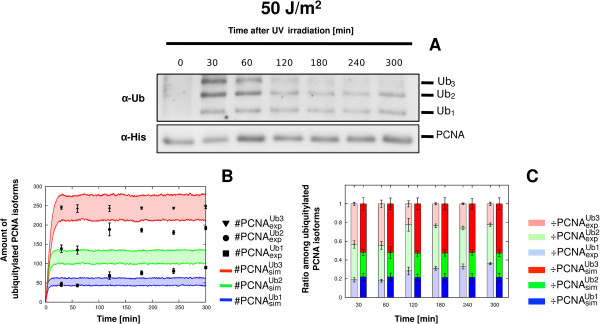
Figure 4.
Comparison between experimental and simulation results of PCNA ubiquitylation dynamics obtained on wild type yeast cells at 50 J/m2 UV dose. The figure shows the experimental measurements on WT yeast cells irradiated at 50 J/m2 UV dose and the comparison with the corresponding simulation results. (A) Representative image of a western blot showing a time-course measurement of mono-, di- and tri-ubiquitylated PCNA isoforms (top part, denoted by α- Ub) and of non modified PCNA (bottom part, denoted by α-His), sampled from 0 to 5 h after UV irradiation. The experiment was repeated 3 times. (B) Comparison between the mean dynamics of mono-, di- and tri-ubiquitylated PCNA isoforms emerging from 100 independent stochastic simulations, and the mean value of experimental data , together with the respective standard deviation . Stochastic simulations were executed starting from the same initial conditions (see Table 2 for molecular amounts and Table 3 for reaction constants) and with an estimated number of DNA lesions equal to 10012. Colored areas indicate the amplitude of stochastic fluctuations around the mean value ). Data are plotted by using the units representation (see Additional file 5). (C) Comparison between the ratio of experimental (, left bars) and simulated (, right bars) ubiquitylated PCNA isoforms at every sampled time point. Mean and standard deviation bars of both experimental and simulated ratios are plotted by using the normalized representation (see Additional file 5).