Table 3.
Citrus grapefruit leaf proteins that were up-accumulated in response to Las-infection
| Spota |
ASVb |
Protein function/namec | Accession # |
Theoreticald |
Se | Mf | Eg | |
|---|---|---|---|---|---|---|---|---|
| UP IP US IS | Mr | pI | ||||||
| |
|
Energy/Metabolisms |
|
|
|
|
|
|
| 29i |
 |
Granule-bound starch synthase |
gi|223029784 |
67320 |
8.56 |
|
|
|
| 33i |
 |
Granule-bound starch synthase |
gi|223029784 |
67320 |
8.56 |
|
|
|
| 61i |
 |
Granule-bound starch synthase |
gi|223029784 |
67320 |
8.56 |
|
|
|
| |
|
Redox homeostasis |
|
|
|
|
|
|
| 34i |
 |
Peroxiredoxins, prx-1, prx-2, prx-3, putative |
gi|255578581 |
29299 |
8.38 |
|
|
|
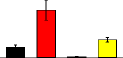
| 119i |
 |
Cu/Zn superoxide dismutase |
gi|2274917 |
12784 |
5.82 |
|
|
|
| 147i |
 |
2Fe-2S ferredoxin-like protein |
gi|18397961 |
17602 |
7.75 |
|
|
|
| |
|
Pathogen response |
|
|
|
|
|
|
| 14i |
 |
Chloroplastic light/drought-induced stress protein |
gi|22261807 |
35216 |
5.24 |
|
|
|
| 15i |
 |
Acidic class I chitinase |
gi|23496445 |
34123 |
4.70 |
|
|
|
| 19 |
 |
Acidic class I chitinase |
gi|23496445 |
36735 |
4.81 |
64 |
8 |
24 |
| 39 |
 |
Lectin-related protein precursor |
gi|11596188 |
29272 |
5.10 |
72 |
7 |
32 |
| 41h |
 |
Chitinase |
gi|1220144 |
32459 |
5.06 |
53 |
5 |
20 |
| 43 |
 |
Chitinase |
gi|1220144 |
36735 |
4.81 |
73 |
11 |
41 |
| 44 |
 |
Lectin-related protein precursor |
gi|11596188 |
29272 |
5.10 |
85 |
7 |
32 |
| 66i |
 |
Chitinase |
gi|1220144 |
31909 |
5.06 |
|
|
|
| 88i |
 |
Chitinase |
gi|1220144 |
31909 |
5.06 |
|
|
|
| 92i |
 |
Chitinase |
gi|1220144 |
31909 |
5.06 |
|
|
|
| 124 |
 |
Chitinase |
gi|1220144 |
36735 |
4.81 |
110 |
11 |
45 |
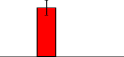
| 141i |
 |
CAP160 protein |
gi|22327778 |
65970 |
5.07 |
|
|
|
| 153 |
 |
Putative miraculin-like protein 2 |
gi|119367468 |
17806 |
6.74 |
105 |
7 |
49 |
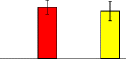
| 156i |
 |
Miraculin-like protein 2 |
gi|11596180 |
25652 |
6.10 |
|
|
|
| 160i |
 |
Chitinase |
gi|1220144 |
31909 |
5.06 |
|
|
|
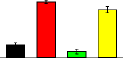
| 179i |
 |
PR-4 type protein |
gi|3511147 |
15227 |
5.50 |
|
|
|
| 181i |
 |
Miraculin-like protein 2 |
gi|87299377 |
24120 |
5.61 |
|
|
|
| 187i |
 |
Miraculin-like protein 2 |
gi|87299377 |
24120 |
5.61 |
|
|
|
| 202 |
 |
Putative miraculin-like protein 2 |
gi|119367468 |
23610 |
8.18 |
126 |
9 |
54 |
| 203i |  |
Miraculin-like protein 2 | gi|87299377 | 24120 | 5.61 | |||
a The spot numbers correspond to the numbers given in Figure 2 and Additional file 3: Figure S2.
b Average spot volume per treatment group; UP, uninfected reference for pre-symptomatic plants; IP, infected pre-symptomatic plants; US, uninfected reference for symptomatic plants; IS, infected symptomatic plants. Average spot volumes separated by letters to show significant difference are presented in Additional file 6: Appendix S1.
c Protein function/name was determined by http://www.ncbi.nlm.nih.gov/BLAST/.
d Theoretical relative molecular weight (Mr) and isoelectric point (pI) were calculated by http://www.expasy.org/. Observed Mr and pI can be extrapolated from Figure 1.
e Mascot score of protein hit.
f Number of matched peptide masses. The sequences of PMF-matched peptides per spot are provided in Additional file 7: Appendix S2.
g Percent sequence coverage of matched peptides.
h Protein identification confirmed by MALDI-TOF-MS/MS. Sequencing information is provided in Additional file 7: Appendix S2.
i Protein identification confirmed by LC-MS/MS.
