Table 1.
Identities of HT induced/decreased proteins in the peels of Satsuma mandarin fruit
| Glycolysis and TCA cycle: 9 | ||||||||
|---|---|---|---|---|---|---|---|---|
|
No. |
Protein name |
Protein accumulation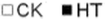 |
Source organism |
GI no. |
TheoMr/pI |
Expt Mr/pI |
PC |
MS |
| H1 |
Enolase |
 |
Glycine max |
42521309 |
47.97/5.31 |
50.84/5.23 |
8 |
443 |
| H2 |
Phosphoglycerate kinase, putative |
 |
Arabidopsis thaliana |
15223484 |
50.02/8.27 |
41.88/4.90 |
9 |
108 |
| H3 |
Putative 2-oxoglutarate dehydrogenase E2 subunit |
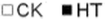 |
Oryza sativa |
48716382 |
49.46/6.78 |
43.64/4.62 |
4 |
105 |
| H4 |
Malate dehydrogenase, cytoplasmic |
 |
Beta vulgaris |
11133601 |
35.81/5.89 |
37.61/4.61 |
7 |
347 |
| H5 |
Triosephosphate isomerase, cytosolic (TIM) |
 |
Zea mays |
136063 |
27.23/5.52 |
24.97/4.47 |
9 |
310 |
| H6 |
Malate dehydrogenase, mitochondrial precursor |
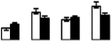 |
Citrullus lanatus |
126896 |
36.40/8.88 |
36.36/4.71 |
8 |
376 |
| H7 |
Pyruvate dehydrogenase E1 beta subunit isoform 2 |
 |
Zea mays |
162458637 |
40.23/5.56 |
35.91/5.72 |
5 |
302 |
| H8 |
NAD-malate dehydrogenase |
 |
Nicotiana tabacum |
5123836 |
43.67/8.03 |
34.10/5.38 |
11 |
436 |
| H9 |
Putative2,3-bisphosphoglycerate- independent phosphoglycerate mutase |
 |
Arabidopsis thaliana |
15982735 |
60.67/5.27 |
61.18/5.12 |
9 |
207 |
|
Redox regulation: 9 | ||||||||
|
No. |
Protein name |
Protein accumulation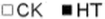 |
Source organism |
GI no. |
TheoMr/pI |
Expt Mr/pI |
PC |
MS |
| H10 |
Isoflavone reductase related protein |
 |
Pyrus communis |
3243234 |
33.80/6.02 |
33.12/4.53 |
5 |
92 |
| H11 |
Oxidoreductase, zinc-binding dehydrogenase family protein |
 |
Arabidopsis thaliana |
15220854 |
41.13/8.46 |
37.58/5.41 |
6 |
93 |
| H12 |
Putative NAD(P)H oxidoreductase, isoflavone reductase |
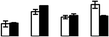 |
Arabidopsis thaliana |
19310585 |
34.24/6.61 |
34.43/4.71 |
4 |
67 |
| H13 |
Aldo/keto reductase family protein |
 |
Arabidopsis thaliana |
42571931 |
27.76/6.33 |
38.36/5.16 |
6 |
164 |
| H14 |
2-oxoacid dehydrogenase family protein |
 |
Arabidopsis thaliana |
15240454 |
50.27/9.19 |
47.44/4.92 |
7 |
168 |
| H15 |
Mitochondrial processing peptidase |
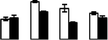 |
Solanum tuberosum |
587562 |
54.62/5.99 |
49.29/5.42 |
4 |
65 |
| H16 |
Copper/zinc superoxide dismutase |
 |
Citrus limon |
33340236 |
15.20/5.46 |
17.02/5.27 |
4 |
263 |
| H17 |
Putative NAD(P)H oxidoreductase, isoflavone reductase |
 |
Arabidopsis thaliana |
19310585 |
34.24/6.61 |
34.47/4.92 |
3 |
85 |
| H18 |
Chloroplast stromal ascorbate peroxidase |
 |
Vigna unguiculata |
45268439 |
39.95/7.06 |
31.00/5.06 |
9 |
191 |
|
Stress response: 7 | ||||||||
|
No. |
Protein name |
Protein accumulation |
Source organism |
GI no. |
TheoMr/pI |
Expt Mr/pI |
PC |
MS |
| H19 |
Abscisic stress ripening-like protein |
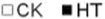 |
Prunus persica |
16588758 |
20.74/5.68 |
29.89/5.25 |
4 |
316 |
| H20 |
Class III chitinase |
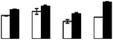 |
Benincasa hispida |
5919201 |
33.03/9.8 |
26.84/6.32 |
3 |
65 |
| H21 |
Heat shock protein 60 |
 |
Prunus dulcis |
24637539 |
58.06/5.26 |
88.79/5.66 |
10 |
205 |
| H22 |
17.7 kDa heat shock protein |
 |
Carica papaya |
37933812 |
17.77/6.39 |
19.12/5.95 |
2 |
98 |
| H23 |
Putative heat shock 70 KD protein, mitochondrial precursor |
 |
Oryza sativa |
27476086 |
70.68/5.45 |
92.99/5.65 |
17 |
358 |
| H24 |
Low molecular weight heat-shock protein |
 |
Pseudotsuga menziesii |
1213116 |
18.18/5.82 |
17.78/6.39 |
4 |
122 |
| H25 |
Beta-1, 3-glucanase |
 |
Citrus sinensis |
2274915 |
37.32/9.19 |
34.38/6.18 |
7 |
98 |
|
Other metabolism: 6 | ||||||||
|
No. |
Protein name |
Protein accumulation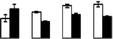 |
Source organism |
GI no. |
TheoMr/pI |
Expt Mr/pI |
PC |
MS |
| H26 |
Fiber annexin |
 |
Gossypium hirsutum |
3493172 |
36.20/6.34 |
36.23/5.53 |
5 |
70 |
| H27 |
Alpha-amylase |
 |
Citrus reticulata |
20336385 |
17.05/4.91 |
44.16/5.64 |
8 |
177 |
| H28 |
Glutamine synthetase |
 |
Elaeagnus umbellata |
47933888 |
39.33/6.10 |
39.38/5.16 |
5 |
134 |
| H29 |
Putative inorganic pyrophosphatase |
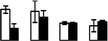 |
Oryza sativa |
46805453 |
20.45/4.61 |
28.58/6.31 |
4 |
74 |
| H30 |
Thiazole biosynthetic enzyme, chloroplast precursor |
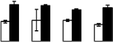 |
Citrus sinensis |
6094476 |
37.74/5.40 |
31.35/6.04 |
12 |
447 |
| H31 |
Lipocalin protein |
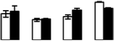 |
Capsicum annuum |
50236424 |
21.41/7.66 |
19.0/4.33 |
3 |
143 |
|
Protein folding: 6 | ||||||||
|
No. |
Protein name |
Protein accumulation |
Source organism |
GI no. |
TheoMr/pI |
Expt Mr/pI |
PC |
MS |
| H32 |
Catalytic/coenzyme binding |
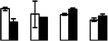 |
Arabidopsis thaliana |
18404496 |
34.97/8.37 |
25.54/4.50 |
6 |
252 |
| H33 |
Chaperonin-60 beta subunit |
 |
Solanum tuberosum |
1762130 |
63.26/5.72 |
57.6/5.77 |
12 |
283 |
| H34 |
Protein disulfide isomerase |
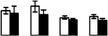 |
Zea mays] |
162461925 |
40.43/6.29 |
37.7/5.26 |
9 |
165 |
| H35 |
Transcription factor APFI |
 |
Arabidopsis thaliana |
13507025 |
30.14/6.24 |
28.15/5.14 |
3 |
89 |
| H36 |
Putative ATP synthase beta subunit |
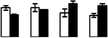 |
Oryza sativa |
56784991 |
45.93/5.33 |
51.13/5.66 |
20 |
695 |
| H37 | Putative ATP synthase beta subunit | Oryza sativa | 56784991 | 45.93/5.33 | 50.0/5.6 | 20 | 808 | |
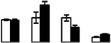
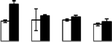
Samples at 1, 6, 12 and 32 DAT were used for differential analysis of proteins accumulation. After 2-DE, protein spot intensities were quantified using PDQuest 2-D software version 7.4. Comparative and statistical analysis shows there were only 50 differentially displayed protein spots whose abundance was altered at least 1.5-fold between the HT and control during the same time period. Of these, 34 proteins were successfully identified using Applied Biosystems 4800 matrix-assisted laser desorption/ionization-time-of-flight tandem mass spectrometry (MALDI-TOF MS) analysis. Protein relative accumulation is represented by the column configuration, and accumulation at 1, 6, 16 and 32 DAT is shown from left to right. Protein name and GI No. are from the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.n ih.gov/guide/). Experimental molecular mass and pI (Exp. M/pI) were calculated using PDQuest software (version 7.4), and mass is shown in kDa. PC: Peptide count. MS: Mascot score.
