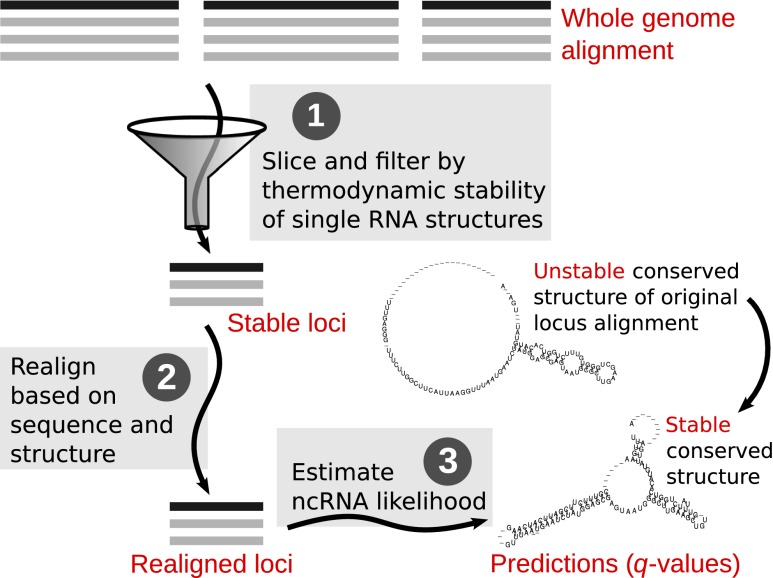
Figure 1.
The whole-genome REAPR (RE-Alignment for Prediction of structural ncRNAs) pipeline. Step 1: The syntenic blocks of the WGA are sliced into windows, which are filtered by thermodynamic stability. Stable windows of the same orientation are merged into stable loci. Step 2: Each candidate locus is realigned based on sequence and structure similarity by LocARNA within limited deviation from the WGA. Step 3: Each realigned locus is evaluated by a de novo ncRNA predictor such as RNAz 2.0 for its likelihood of containing structural ncRNA. The evaluation is then transformed into a q-value to control FDR.