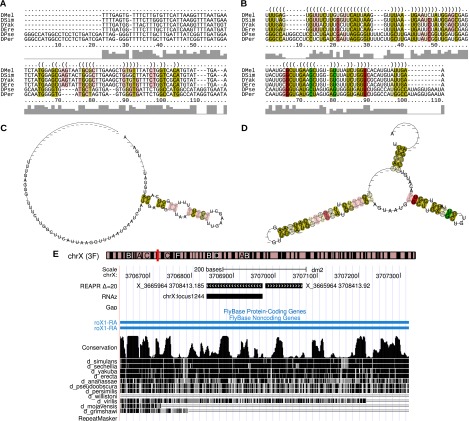
Figure 6.
A novel potential structural motif in the long ncRNA roX1 of the D. melanogaster chromosome X. The alignment is located on columns 9363200–9368080 in the syntenic block X_3665964_3708413 of the fly WGA and corresponds to positions 3706976–3707066 of the D. melanogaster chromosome X. The figure shows the locus alignment of the original WGA (A) and that realigned with LocARNA Δ = 20 (B). RNAalifold (Bernhart et al. 2008) was used to compute the consensus secondary structures (C,D) inferred from A and B, respectively, and generate the graphics of figures (A–D). The colors indicate the structure conservation (saturation) and number of compensatory mutations (hue) in base-paired alignment columns. (E) UCSC Genome Browser visualization of the location of this prediction (X_3665964_3708413.92) and a neighboring one (X_3665964_3708413.185).