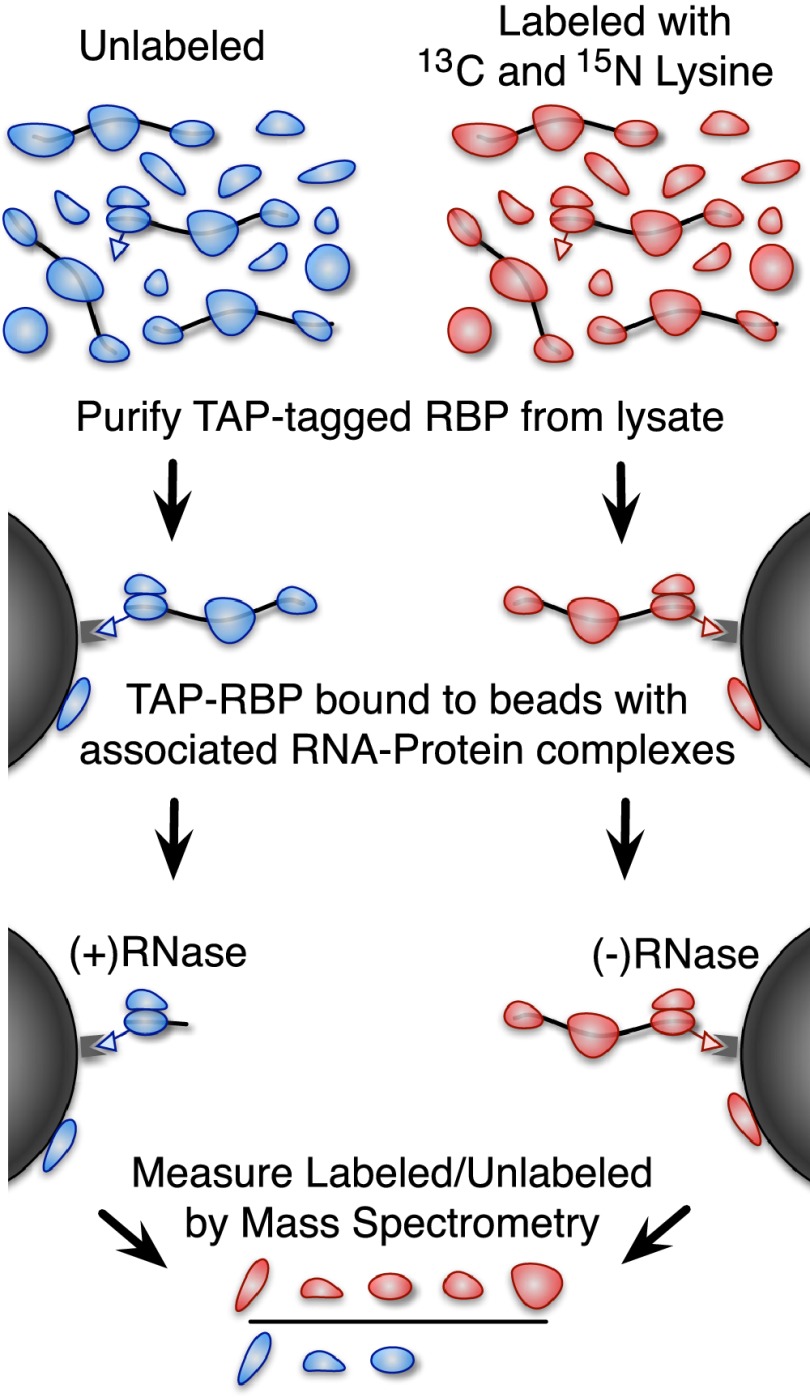
Figure 1.
A method for identifying RNA-dependent protein interactions. An overview of our proteomic method for identifying RNA-dependent protein interactions. A yeast strain with a TAP-tagged protein of interest is grown in media labeled with heavy (13C and 15N isotope enriched) lysine or unlabeled media. The cells are lysed and the protein of interest is purified using the TAP tag. The unlabeled sample is treated with RNase, and the heavy labeled sample is not. The beads are then boiled in SDS-PAGE buffer to release any bound proteins and combined as heavy labeled RNase untreated and unlabeled treated with RNase (we also performed the inverse as a replicate and to control for labeling-related artifacts). The heavy-to-light ratio measured by mass spectrometry indicates the fraction of the bound protein that was liberated by RNase treatment.