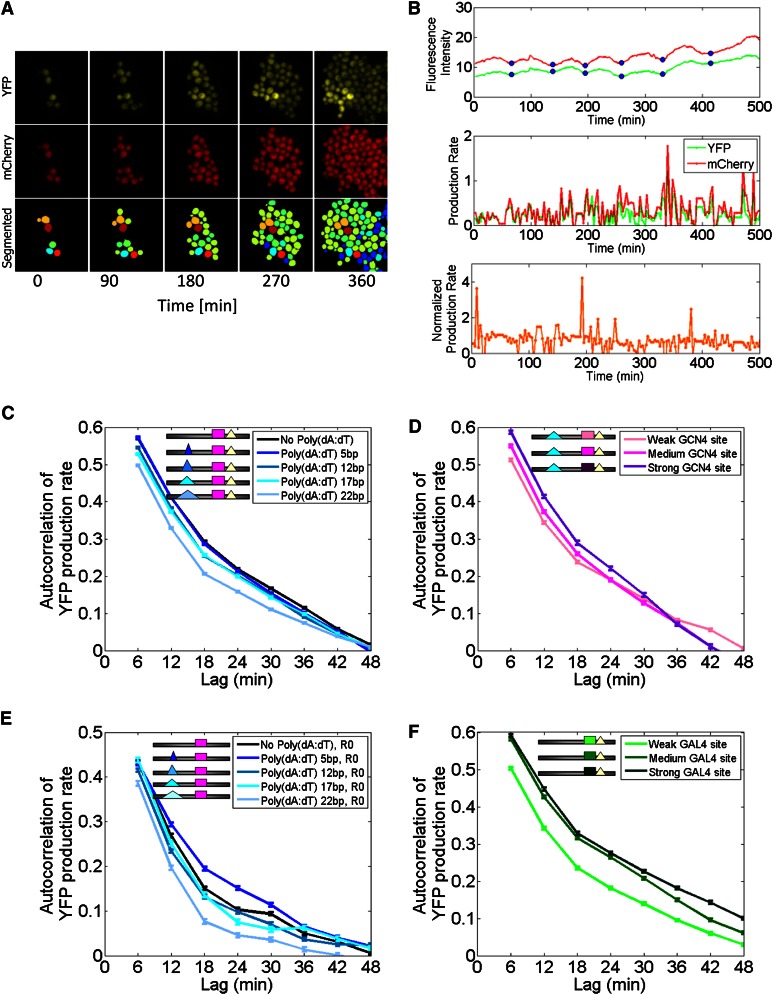
Figure 2.
Opposing effects on promoter dynamics for lengthening poly(dA:dT) tracts and strengthening transcription factor binding sites. (A) Representative time-lapse microscopy images of one imaging area at five different time points, displaying YFP fluorescence (top), mCherry fluorescence (middle), and automatically segmented cells. (B) Representative time-lapse traces of YFP and mCherry fluorescence of a single cell over time (top), along with YFP and mCherry production rates (middle), and normalized YFP production rates (bottom; normalization done by mCherry; see Methods). Blue circles denote cell cycles. (C) Longer poly(dA:dT) tracts result in faster promoter dynamics. Shown is the average autocorrelation of normalized YFP production rates across thousands of different cell traces for each of five different promoter variants with poly(dA:dT) tracts of length 0, 5, 12, 17, or 22 bp. Bars denote standard error. (D) Higher-affinity binding sites result in lower promoter dynamics. Same as C, for three promoter variants that differ only in the affinity of the Gcn4 site. (E) Same as C, but where the poly(dA:dT) tract variants were inserted into a different genetic background in which the right poly(dA:dT) tract is deleted (R0). (F) Same as D, but for three Gal4 sites that differ in their affinity.