Fig. 1.
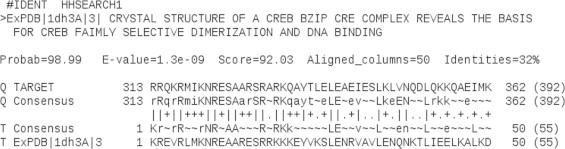
Pairwise sequence alignment between the bZIP regions of query (ABF1) and template (CREB) as obtained from HHsearch method of SWISS-MODEL template identification tool. The first 50 residues of ABF1 bZIP region (313–362) and CREB bZIP (1–50) were aligned with sequence identity 32%, E-value of 1.3e−09 with similarity score 92.03. Consensus (Q for query and T for template) is defined on the basis of profile HMMs of respective sequences where the one letter codes define the most probable amino acid residues observed at the respective positions (capital letters denote a probability of occurrence which is greater than or equal to 60%, small letters if it is greater than or equal to 40% and tilde (∼) denotes non-conserved positions).
