Fig. 1.
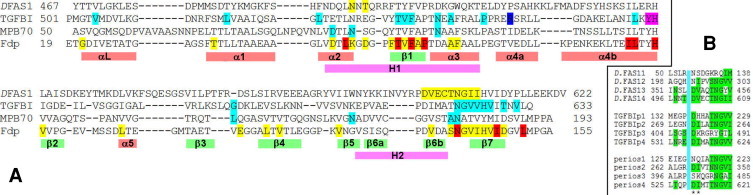
Sequence alignments of FAS1 domains. (A) Alignments based on structural similarity. Sequences are those of the four FAS1 domains with three-dimensional structures: The Drosophila FAS1-3/4 pair (DFAS1: PDB 1o70, domain 4); the fourth FAS1 domain of human TGFBIp (TGFBI: PDB 2vxp); the M. bovis secreted protein MPB70 (MPB70: PDB 1nyo); and Fdp (Fdp: PDB 1w7d). The NMR structure of the fourth FAS1 domain of human TGFBIp (PDB 1x3b) is very similar to the crystal structure and was not used as an independent structure. Colour code: in DFAS1, yellow denotes residues described here as being interacting residues. In TGFBIp, blue denotes R555, one of the two major sites giving rise to corneal dystrophy. Other disease-causing sites are indicated in cyan. The sequence YH, suggested as a possible binding site [16], is shown in magenta. In MPB70, cyan indicates suggested interaction sites [32]. Highly conserved and completely conserved residues are indicated on Fdp in yellow and red respectively. Locations of regular secondary structure, and the conserved regions H1 and H2, are indicated below the sequences. (B) Domains 1 through 4 from Drosophila FAS1, TGFBIp and periostin, each of which contains four tandem FAS1 domains. The alignments encompass the two regions (separated by a blue box) discussed here as being binding sites. Comparisons are more reliable in the second sequence, which is longer and better conserved. Conserved residues are highlighted in green; the important DI/V sequence is marked by asterisks. Domains 2 and 4 are more highly conserved. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
