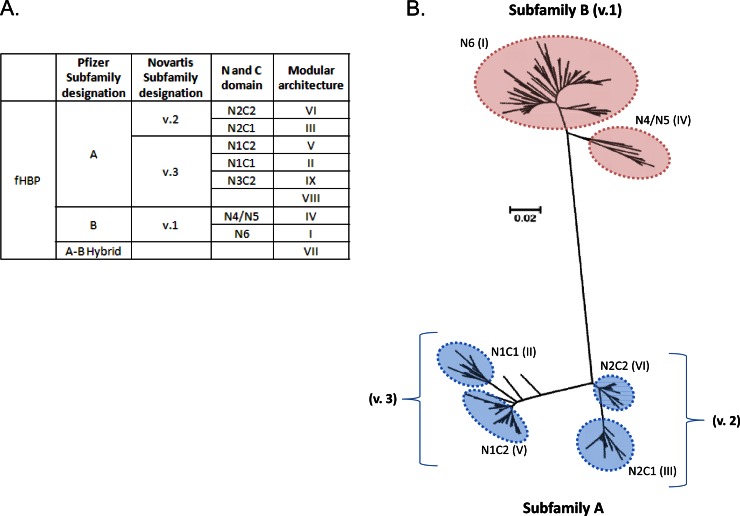
Fig 2.
Phylogenetic analysis of fHBP protein sequences. (A) Nomenclature used to describe fHBP, including Pfizer and Novartis subfamily designations, N- and C-terminal domains, and Pajon modular architecture assignations (20, 21, 29, 31). (B) Neighbor-joining tree of 569 fHBP variants from http://pubmlst.org/neisseria/fHbp/. The tree was generated in ClustalW (145) with 500 bootstraps and drawn using MEGA 5.05 (146). Subfamily designations are defined by Murphy et al. and Masignani et al. (20, 29). Subfamily A (variants 2 and 3) can be further subdivided into 4 major groupings based on the N- and C-terminal domains of fHBP (N1C1, N2C2, N1C2, and N2C1), and subfamily B can be divided into 3 groupings (N4, N5, and N6). Pajon et al. have made similar groupings based on five variable segments, dividing all fHBP variants into 9 modular groups (31). The six major modular groups are indicated by the roman numerals (the minor groups VIII and IX, each represented by a single variant, are not labeled on the tree). Not shown are several hybrid sequences that are composed of both subfamily A and subfamily B regions. N2C1 and N2C2 together are equivalent to variant 2 (v.2), and N1C2 and N1C1 are equivalent to variant 3 (v.3). The bar indicates genetic distance.