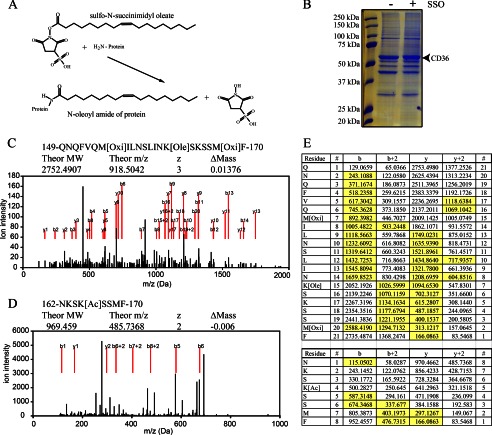
FIGURE 1.
Identification of SSO-binding site in CD36. A, schematic representation of the SSO protein labeling reaction showing that when the sulfo-N-succinimidyl oleate ester reacts with a primary amine group on a membrane protein, an N-oleoyl amide of the target protein is formed together with the NHS group. B, Coomassie-stained gel showing the deglycosylated CD36 bands at about 53 kDa. CHO cells stably expressing wild type FLAG-tagged CD36 (CD36WT) were used. The cells were labeled with 100 μm SSO as indicated, and the CD36 protein was immunoprecipitated, deglycosylated, and resolved on a gel. The CD36 bands were extracted and subjected to mass spectrometry. C, mass spectrometry evidence of oleoylation of lysine 164. CD36 protein chymotryptic fragments were ionized into y, y2+, b, and b2+ series with m/z values detected by MS. y and b series are marked with red lines. Modification by SSO (Ole) was detected together with oxidation of methionines (Oxi), caused by ionization. Spectrum of oleoylated peptide is shown. D, mass spectrometry evidence of acetylation (Ac) of lysine 166, details as above. E, sequence of the fragments and complete b and y series ions generated by the ionization. Oleoylation of Lys-164, top table; acetylation of Lys-166, bottom table. Detected and assigned ions are highlighted in yellow.