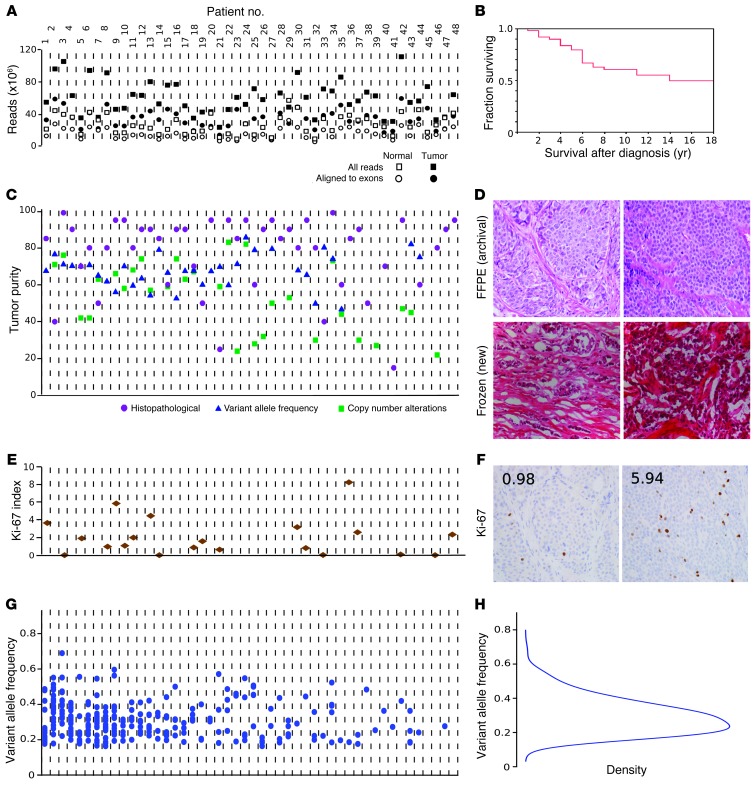
Figure 1. Tumor and sequencing characteristics.
Tumor-associated (somatic) mutations in the exome were determined by nextgen sequencing. (A) Sequencing quality control. Total raw sequencing reads and enrichment for the targeted exome regions were similar across samples. Quality control measures were uncorrelated with downstream results, supporting that observed disparities among tumors represent biological diversity of SI-NETs. x axes in A, C, E, and G correspond to individual tumors. (B) Survival of 48 patients. See also Supplemental Figure 2. (C) Tumor cell purity was determined by histopathology, VAF (fraction of mutated sequencing reads), and copy number alteration estimates (based on the ratio of sequence read counts for a tumor-deleted chromosome compared with germline). The 3 methods cross-validated the observation that mean tumor content was high across the dataset and uncorrelated with biological results. (D) Formalin-fixed, paraffin-embedded (FFPE) and frozen sections of 2 cases, demonstrating tumor purity and well-differentiated histology. (E) Ki-67 labeling index, demonstrating that the majority of cases were WHO 2010 classification low grade (≤2%), and a minority were intermediate grade (>3%). See also Supplemental Figure 3. (F) Ki-67 immunohistochemistry for cases in D. Numerals indicate the Ki-67 index. (G) Mutation counts and VAF for individual tumors. The number of mutations is shown for each patient as a scatter plot of the VAF for each event. While the sensitivity of sequencing technology detecting alterations in subsets of tumor cells is limited by sequence read depth, the distribution of VAF suggests that the reported mutations are dominant throughout the entirety of each tumor. (H) Unimodal distribution of VAF among mutation calls. Original magnification, ×40.