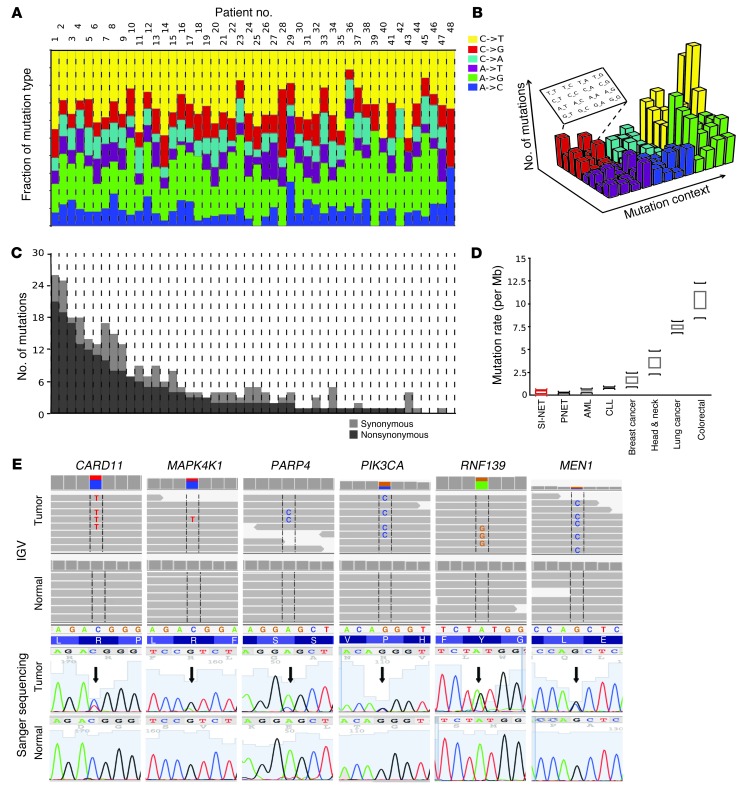
Figure 2. Somatic mutation landscape across the coding genes (exome) in SI-NETs.
(A) Mutation pattern. A similar characteristic nucleotide mutation pattern was observed in all tumors. Somatic SNVs were most commonly the result of either of the 2 possible “transitions”: C>T (pyrimidine-to-pyrimidine) or A>G (purine-to-purine). The 4 possible transversions (pyrimidine-to-purine or reverse) were uncommon. x axes in A and C correspond to individual tumors. (B) Sequence context of SNVs. Extending the analysis of mutation patterns to the 16 possible motifs of nucleotides preceding and following an SNV, a pattern corresponding to known mechanisms of mutagenesis was observed. Most prominent was the CpG context, rendering the base C susceptible to deamination resulting in uracil and subsequent replacement by thymidine. Figure adapted from Journal of Clinical Investigation (29). (C) Mutation rate. SI-NET exonic mutation counts were uniform among tumors varying within only 1 order of log10 magnitude, suggesting absence of a hypermutator subtype (usually seen with mismatch repair deficiencies). (D) Comparison of mutation rate with well-characterized human cancers. Shown is the median mutation rate and range reported for the indicated cancer types. (E) Somatic SNVs representative of the preponderance of mutations in cancer genes in SI-NETs. Top: Primary sequence data showed each respective mutation in a fraction of the sequence reads consistent with an alteration in only 1 gene allele in the tumor or contamination of the tumor sample with normal tissue. No mutation was seen in any sequence read of the germline. SNVs are framed and shown as letters within the reads. IGV, Integrative Genomics Viewer. Bottom: Sanger sequencing. Arrows denote SNVs. PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit α.