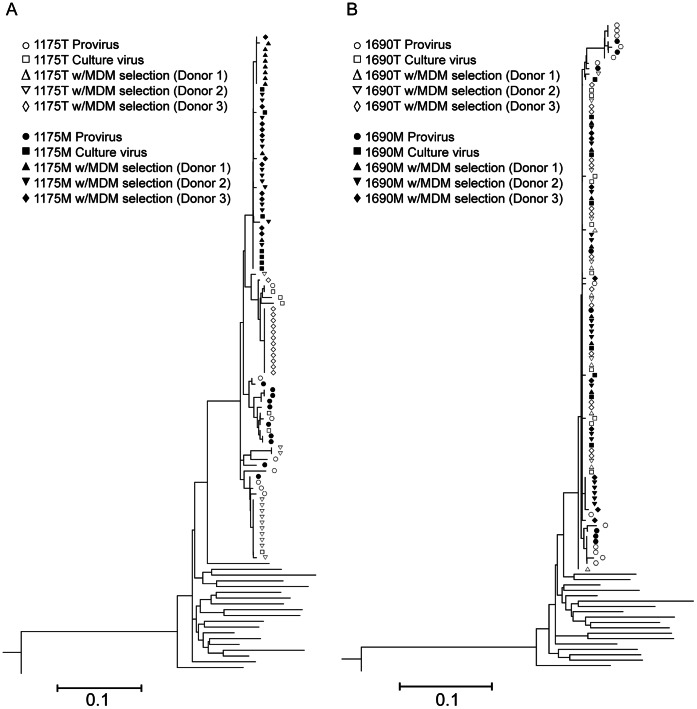
Figure 3. Phylogenetic analysis of HIV-1 env C2-V3-C3 sequences of monocyte- and CD4+ T cell-derived HIV-1.
The neighbor-joining trees of 1175 M and 1175 T (A) as well as 1690 M and 1690 T (B) were shown, respectively. The env C2-V3-C3 sequences of provirues as well as primary isolate before and after MDM selection were acquired from limiting dilution PCR. MDM selection experiments were using seven-day old MDM prepared from 3 healthy donors, respectively, at MOI = 0.01. On Day 22 post-infection, supernatants of infected MDM were subjected to sequence analysis. The viral nucleotide sequences were used to construct neighbor-joining trees based on nucleotide pairwise distances. Reference sequences (not labeled) are HIV-1 subtype B sequences from GenBank database (http://www.ncbi.nlm.nih.gov/).