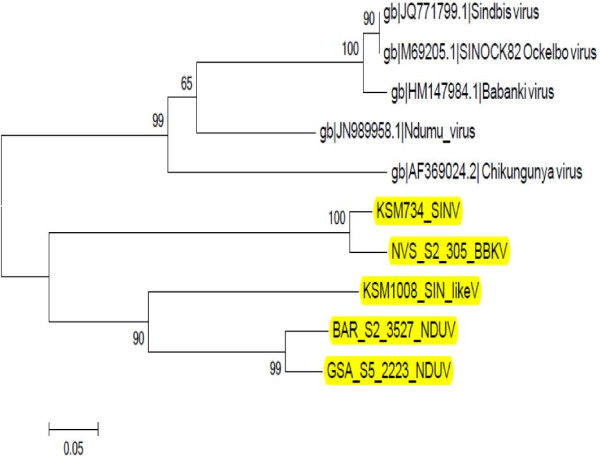
Figure 1.
Neighbour joining phylogenetic tree of nucleotide sequences of selected Alphavirus isolates and reference sequences. The sequences of the isolates reported in this paper (highlighted in yellow) were compared to other members of the alphavirus genus from Genbank [GenBank: JN989958.1, GenBank: JQ771799.1, GenBank: M69205.1, GenBank: HM147984.1, GenBank: AF369024.2]. The Non structural Protein 4 (NSP4) coding regions of these nucleotide sequences, and those of the selected Alphaviruses in this study were aligned using clustalW and the phylogenetic tree constructed using MEGA v5.05. Numbers on internal branches indicate bootstrap values for 1000 replicates. The results show that the Alphaviruses have grouped into two with one group having the isolates under study (from Kenya) and the other has the reference sequences (from other countries). Within the Kenyan group, the Ndumu virus isolates have clustered separately from the Sindbis and the Sindbis-like viruses. This is due to the diverse nature of Alphaviruses. GSA: Garissa; KSM: Kisumu; NVS: Naivasha; BAR: Baringo. NDUV: Ndumu virus; SINV: Sindbis virus; BBKV: Babanki virus.