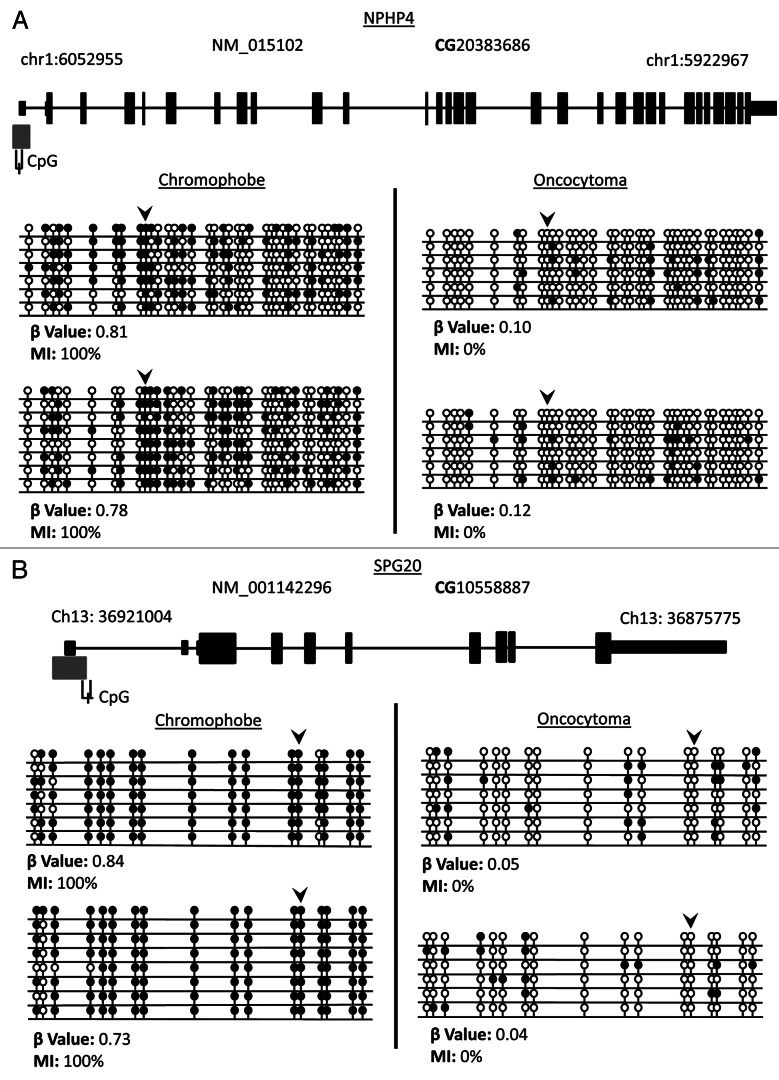
Figure 6. Validation of loci β values using bisulfite clone sequencing of NPHP4 and SPG20 genes. (A) Schematic overview of the NPHP4 gene (black boxes represent exons, black lines represent introns and gray boxes represent CpG islands). Black line below CpG island represents region interrogated by primers for bisulfite sequencing including differentially methylated CpG locus (black dash). The differentially methylated CpG resides in a CpG island within 200 bp of the TSS of the gene. (B) Schematic overview of the SPG20 gene. The differentially methylated CpG resides in a north shore in the 5′UTR region of the gene. Bisulfite sequencing rows represent individual alleles, with each circle indicating location and methylation state of a CpG locus (black circle: methylated; white circle: unmethylated). Black arrowheads point to the CpG loci identified on the 450K array as showing significant differential methylation. Bisulfite sequencing of chromophobe RCC and renal oncocytoma samples confirm that the CpG loci in NPHP4 and SPG20 are hypermethylated in chromophobe RCC samples and unmethylated in renal oncocytoma samples.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.