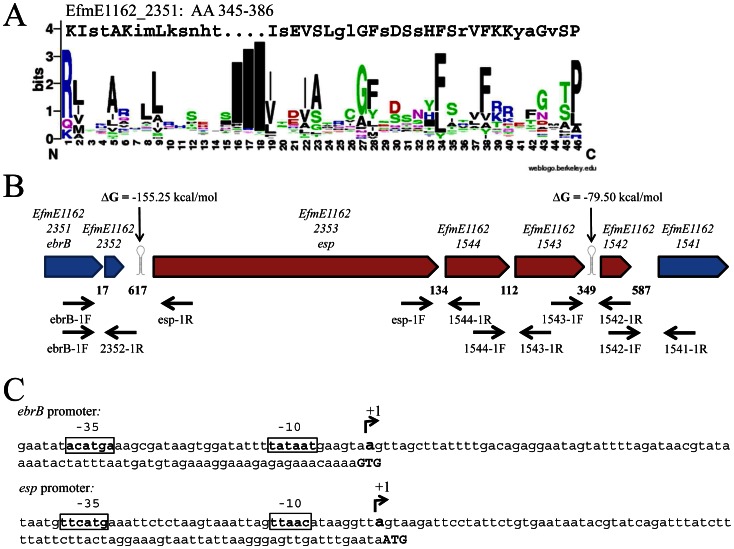
Figure 1. Structural organization of the ebrB region.
(A) Alignment of the putative helix-turn-helix motif, amino acids 345-386, of EbrB to a sequence logo generated using a Prosite database containing 310 HTH motifs (accession number PS00041) in Weblogo. Conserved amino acids are depicted in capital letter. (B) Overview ebrB region, including the esp operon in red, primer sites and predicted transcription terminators (RNAfold). Numbers indicate the intergenic distances in bp (C) ebrB and esp promoter mapping, in bold/box putative -35 and -10 sequence and transcription start (+1).