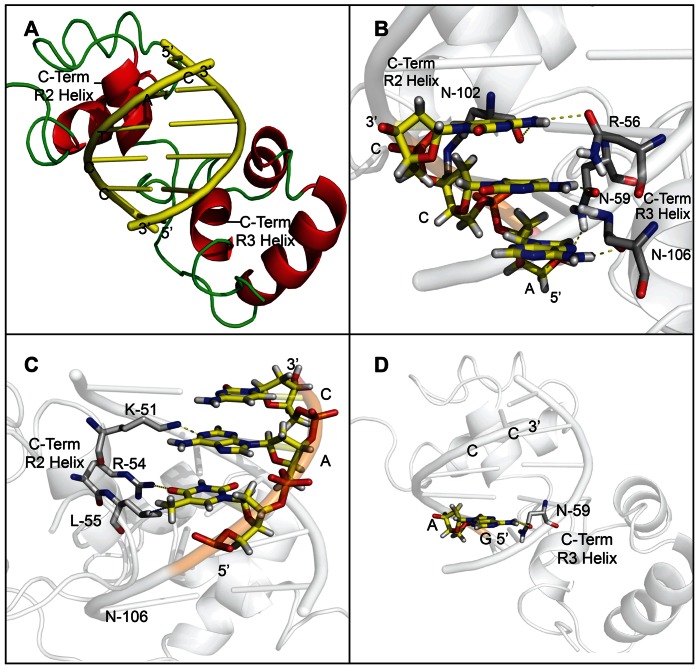
Figure 3. Molecular modelling of AtMYB61 with target sequences confirm binding preferences determined by nitrocellulose filter-binding assays and EMSAs.
(A) Pymol models of ACCTAC motif docked into the binding site of AtMYB61. Molecular modelling was completed by using the online program PHYRE (Protein Homology/analogY Recognition Engine) to predict a crystal structure of AtMYB61 using homology to the c-MYB DNA binding domain. The PBD (protein data bank) file recovered from the PHYRE analysis was used to superimpose the predicted AtMYB61 crystal structure with the c-MYB crystal structure using DaliLite. Using Pymol the 3D sequence model – ACCTAC – was docked into the predicted binding sites of AtMYB61. The AC1 element model is displayed in yellow, the loop secondary structure of AtMYB61 inferred model is displayed in green, and the helix secondary structure of AtMYB61 inferred model is displayed in red. (B) Model of AtMYB61 binding site with the first three ACC nucleotides in the ACCTAC sequence determines that these nucleotides are essential for binding. The AC1 (ACCTAC) nucleotide model was docked into the predicted binding site of AtMYB61. The specific hydrogen bonding between the amino acids of AtMYB61 binding site to the ACC nucleotides of AC1 were predicted by Pymol and listed as follows: asparagine-59 (R3 helix) hydrogen to adenine-1 nitrogen; asparagine-106 (R3 helix) oxygen to adenine-1 hydrogen; asparagine-59 (R3 helix) oxygen to cytosine-2 hydrogen; asparagine-102 (R3 helix) oxygen to cytosine-3 hydrogen; and arginine-56 (R2 helix) oxygen to cytosine-3 hydrogen. This confirms binding data determined by the nitrocellulose filter-binding assay and EMSAs, iterating that the ACC motif is the core recognition motif of AtMYB61. (C) Model of AtMYB61 binding site with the TAC nucleotides in the ACCTAC sequence determine that these nucleotides are less essential for binding. The AC1 (ACCTAC) nucleotide models were docked into the predicted binding sites of AtMYB61. The molecular interactions between the amino acids of AtMYB61 binding site and the TAC nucleotides of AC1 were analyzed by Pymol and are listed as follows: leucine-55 (R2 helix) methyl group was predicted to form a non-polar bond with thymidine-4 methyl group; Arginine-54 (R2 helix) hydrogen was predicted to form a hydrogen bound with thymidine-4 oxygen; lysine-51 (R2 helix) hydrogen was predicted to form a hydrogen bound with adenine-5 nitrogen; and cytosine-6 remained unbound in the model. (D) Model of AtMYB61 binding site with non-binding site (GAGACC) predicts that this motif is not recognised by AtMYB61. The non binding site model was docked into AtMYB61 binding site via Pymol and hydrogen bonding was analyzed. Only one hydrogen bond was predicted between AtMYB61 asparagine-59 (R3 helix) oxygen and the non-binding site adenine-2 hydrogen. Yellow dashed lines indicate hydrogen bonding established by Pymol program, and blue dashed lines indicate non-polar interactions.