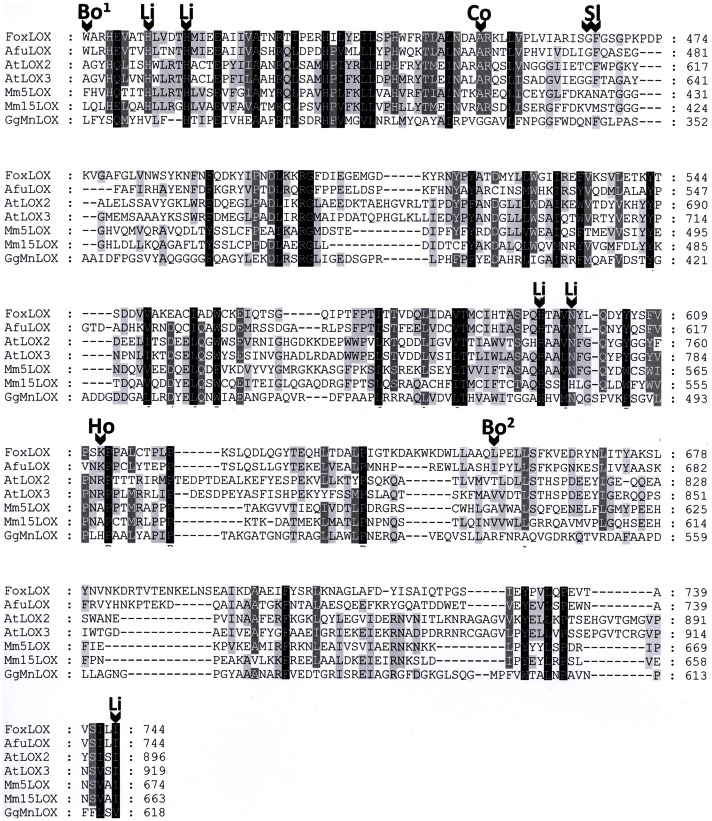
Figure 2. Partial amino acid alignment of different LOX forms with FoxLOX.
Sequences were chosen from F. oxysporum (Fox), A. fumigatus (Af), G. graminis (Gg), A. thaliana (At) and M. musculus (Mm). Alignments were performed using the ClustalX software package employing default parameters. Different determinants for regio- and stereo specificity are indicated as “Bo” (Borngräber according to [66]), “Ho” (Hornung according to [44]), “Sl” (Sloane according to [43]) and “Co” (Coffa according to [45]). Amino acid residues involved in iron binding are indicated by “Li”.