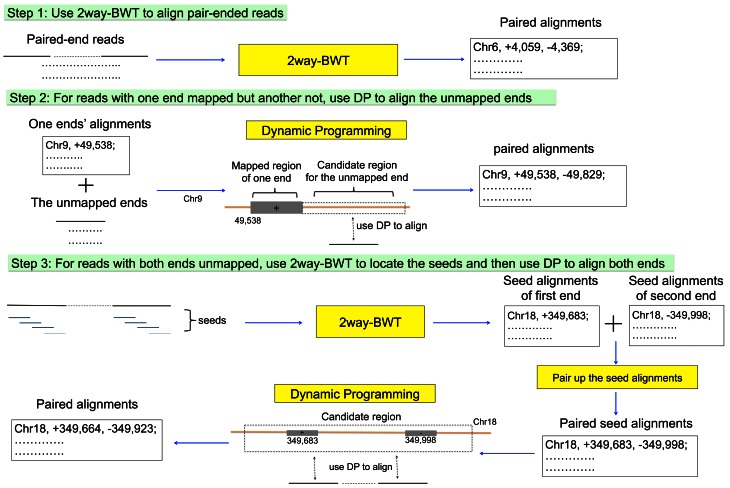
Figure 1. Alignment workflow.
For each read (paired-end specifically, single-end is only with step 1 and step 3), the alignment would be decided in at most three steps. In step 1, SOAP3-dp aligns both ends of a read-pair to the reference genome by using GPU version 2way-BWT algorithm (Methods). Pairs with only one end aligned proceed to step 2 for a GPU accelerated dynamic programming (Methods) alignment at candidate regions inferred from the aligned end. Pairs with both ends unaligned in step 1 and those ends failed in step 2 proceed to step 3 to perform a more comprehensive alignment across the whole genome until all seed hits (substrings from the read) are examined or until a sufficient number of alignments are examined.