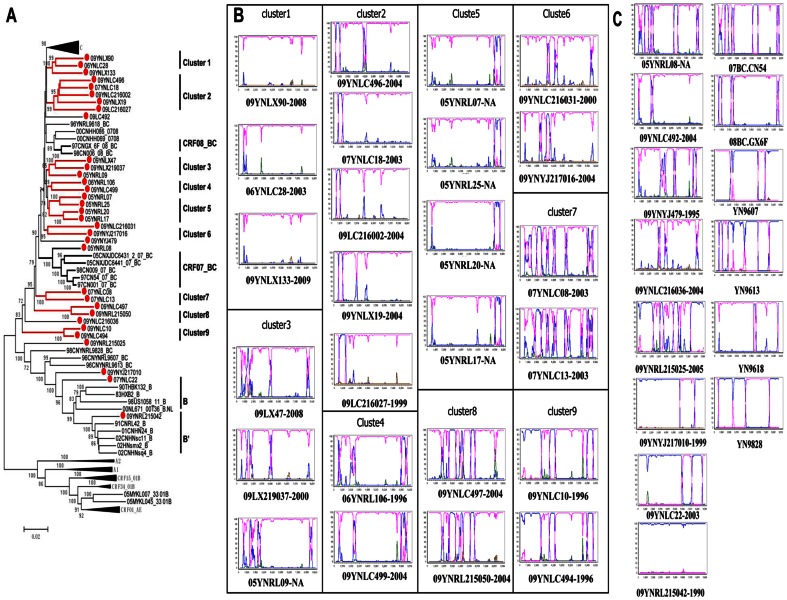
Figure 2. Phylogenetic tree and the boot-scanning plots of near-full-length sequences of HIV-1 strains isolated from Dehong.
(A) The phylogenetic tree of 33 near-full-length sequences of from Dehong IDUs was constructed with MEGA 5.0 using the neighbor-joining method. The subtype reference sequences from the Los Alamos HIV Sequence Database were included as the reference sequences. The stability of the nodes was assessed by bootstrap analyses with1,000 replications. Only bootstrap values of more than 70 are shown at the corresponding nodes. The sequences from IDUs in Dehong are labeled by solid red circles. The nine clusters are shown in red lines. (B)The bootscanning plots of 25 near-full-length sequences in the nine clusters.(C)The bootscanning plots of 8 near-full-length sequences outside the clusters and 6 previously published near full-length sequences of B′/C recombinants in China. In the recombination analyses, a subtype C strain from India (95IN21068) ( pink) and a subtype B′ strain from Yunnan (RL42) (blue) were used as subtype references, and a CRF01_AE strain from Thailand (CM240) (green) was used as an out-group control. In (B) and (C), the number after the sequences ID showed the year of diagnosis of each case.