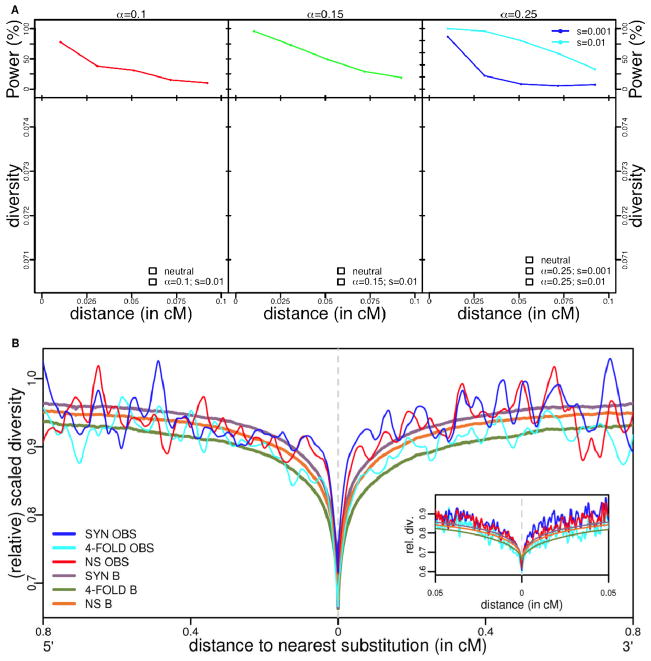
Figure 3.
A. The power to detect a decrease in diversity levels around amino acid substitutions due to classic sweeps. The top panel presents the power at a given genetic distance from the substitution, for the three sets of selection parameters (see (20)). The bottom panel shows the diversity patterns expected around amino acid substitutions for four sets of selection parameters, as well as for a model of purely neutral fixations, after LOESS smoothing (with a span of 0.2). For each set of parameters, the shaded area represents the central 95%-tile obtained from 100 bootstrap simulations. The depth of the trough reflects the fraction of substitutions that were beneficial and its width the typical strength of selection (24). B. Relative diversity levels around non-synonymous, synonymous and four fold degenerate synonymous substitutions predicted under a model of background selection(see (20)). B is the predicted diversity level relative to what is expected with no effects of background selection, i.e., under strict neutrality, taking into account variation in mutation rates (11). OBS is the observed value of average scaled diversity (i.e., diversity divided by divergence to rhesus macaque). For the expected diversity around exons and CNCs, as well as predictions for the X chromosome, see Fig. S8.