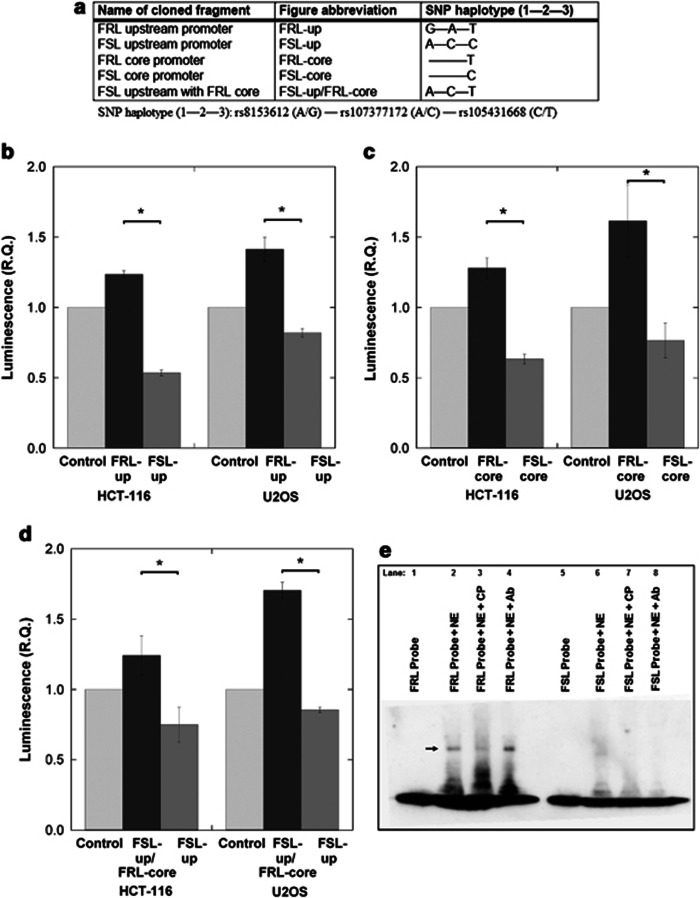
Figure 3.
Luciferase reporter assays and electrophoretic mobility shift assay (EMSAs) were performed to investigate the in vitro transcriptional functionality of the three single-nucleotide polymorphism (SNPs) located in the neuropeptide Y (Npy) promoter region of Flinders resistant line (FRL)/Flinders sensitive line (FSL) rats and the putative DNA–protein interactions at the rs105431668 locus, respectively. (a) Luciferase reporter constructs included either the upstream Npy promoter region (containing all three SNPs: 5′- rs8153612— rs107377172—rs105431668-3′ in a wild-type or a chimeric (FSL upstream with FRL core) form) or the core promoter region (containing only rs105431668). (b, c, d) Collectively, by using two different cell lines (HCT-116 and U2OS), the different luciferase constructs demonstrated that the rs105431668's T-allele (present in homozygosity only in the FRL rats) leads to increased transcription in vitro compared with the C-allele (present only in the FSL rats). (e) Lanes one and five of the mobility shift assays mark the positions of the free (non-interacting) FRL and FSL probes, respectively. Lane two demonstrates an in vitro DNA–protein interaction after the addition of nuclear extract, which is shown as a gel-shift next to the arrow. A strong shift was only generated by the FRL probe (containing the T-allele of rs105431668; lane 2) and not by the FSL (C-allele) probe (lane 6). Lane three shows the reduction of the DNA–protein interaction that occurs after the addition of ‘cold' (unlabeled) probe and indicates specificity in the type of DNA–protein interaction between FRL probe and components of the nuclear extract. Lanes 4 and 8 test for putative supershifts by the addition of an antibody (Ab) against CREB1. No supershifts were observed indicating that CREB1 isn't responsible for the in vitro DNA–protein interaction. Convincingly, however, the initial gel shift is reestablished at the same position for FRL (lane 4) and, again, no shift is observed for FSL (lane 8). *P<0.05.