Figure 1.
Inhibition of p110β by p85-Type Regulatory Subunits and Activation of the Complexes by RTK Phosphopeptide
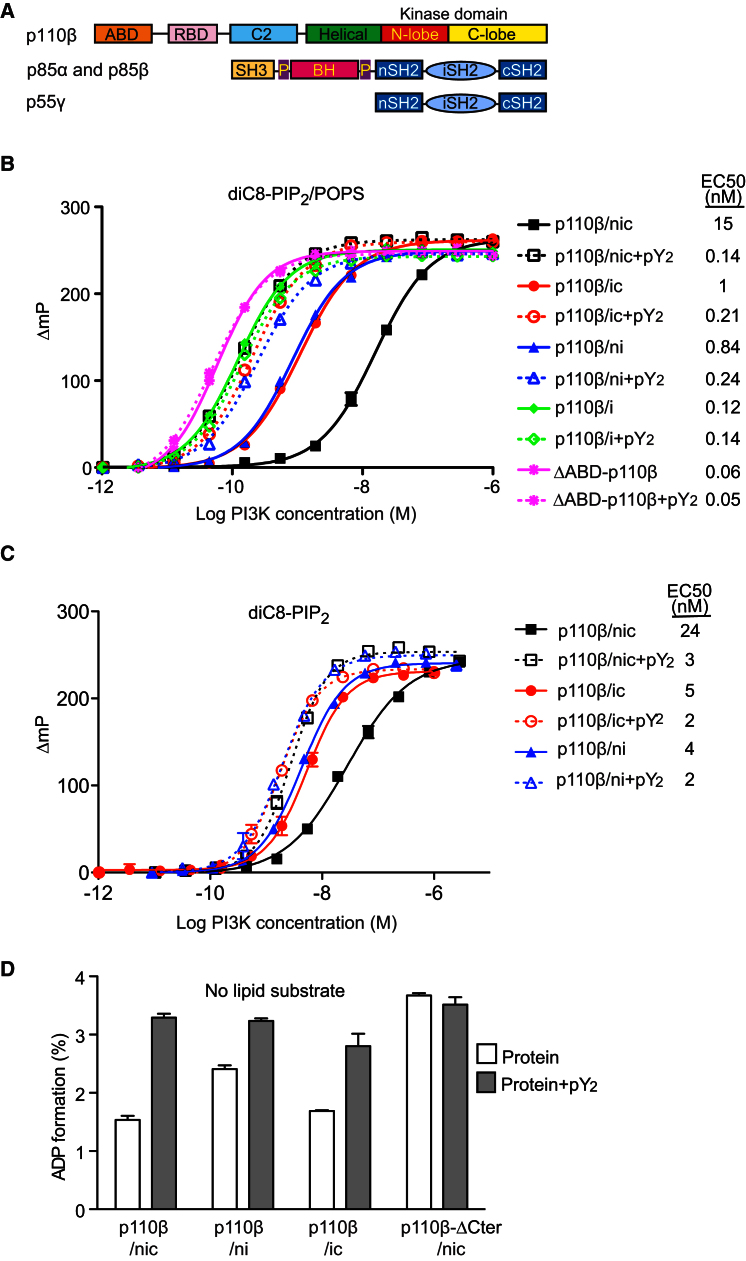
(A) Domain organization of p110β and the regulatory subunits p85α, p85β, and p55γ. The color scheme for the domains is used for all figures, unless otherwise stated.
(B) Kinase activity with diC8-PIP2/POPS liposomes as a function of enzyme concentration (measured by ADP formation) shows the inhibitory effects of p85β-nicSH2 (nic) and p85β-icSH2 (ic) on the basal activity of p110β. The inhibition is released upon addition of the 10 μM PDGFR pY2. The free catalytic subunit (ΔABD-p110β) is more active than any complex. The y axis is expressed as a change in fluorescence polarization (ΔmP), which is obtained by subtracting the observed polarization for the construct at a given enzyme concentration from the maximum fluorescence polarization for that construct. The plateau in these assays arises due to the competitive nature of the ADP detection system, based on displacement of ADP-Alexa 633 tracer from the ADP2-antibody by ADP generated during the PI3K assay (see Supplemental Experimental Procedures). All measurements were done in triplicates, and the error bars indicate the standard error of the mean (SEM).
(C) Kinase activity of p110β/p85β complexes with monomeric substrate (75 μM diC8-PIP2) in the presence and absence of pY2; y axis as in (B).
(D) Basal- and pY2-stimulated activities of 35 nM PI3K complexes in the absence of lipids (ATP hydrolysis). Activities are given as percent of ATP converted to ADP using the Transcreener assay. Bars indicate SEM.