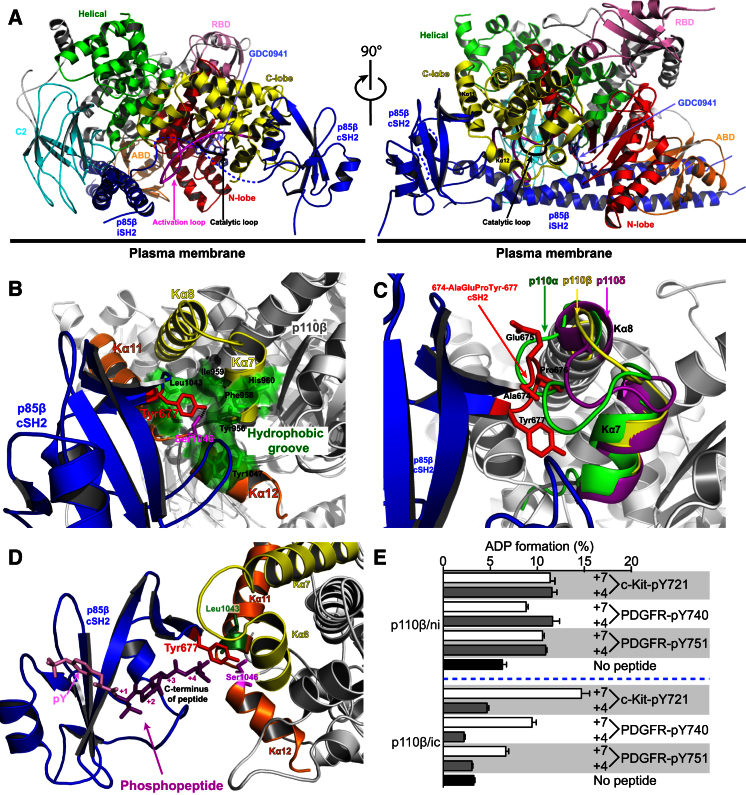
Figure 2.
Structure of p110β in Complex with p85β-icSH2
(A) Cartoon representation of the p110β/p85β-icSH2 complex with GDC0941 (light blue sticks). Views from the front and side (turned 90°) are shown. The ordered activation loop is colored in magenta and the catalytic loop in black.
(B) Detailed view of the contacts between the p85β-cSH2 and the C-lobe of the kinase domain. The main contact residue Tyr677-p85β (red) interacts with Ser1046-p110β (magenta) and the hydrophobic groove (pale green) formed by two “arms,” Kα7/Kα8 (yellow) and Kα11/Kα12 (orange). Dashed black line is a potential hydrogen bond between Tyr677-p85β and Ser1046-p110β.
(C) Cartoon of the p110β Kα7/Kα8 “arm” (yellow) superimposed on p110α (green) and p110δ (magenta), suggesting a steric clash between cSH2 protrusion (674-AlaGluPro-676) (red stick) and Kα7/Kα8 loop of p110α.
(D) Cartoon of the cSH2 binding to RTK phosphopeptide, modeled as in the crystal structure of free cSH2 in complex with a PDGFR phosphopeptide (magenta; PDB ID: 1H9O).
(E) Effects of different RTK phosphopeptides (10 μM) on activity of niSH2 and icSH2 complexes with p110β (1 nM), showing selective requirement for peptides longer than pY + 4 for disinhibition of the cSH2. Error bars indicate SEM.