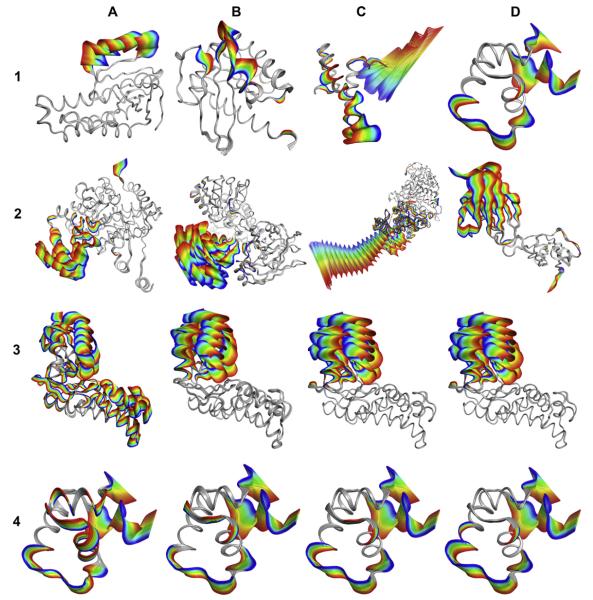
Fig. 3.
(1–2) Examples of protein conformational changes shown as ViewMotions Rainbow diagrams. The Database of Molecular Movements [1] defines three classes of molecular motions two of which are depicted here. (1) Type I corresponds to motions of fragments smaller than domains, and (2) Type II corresponds to domain motions. Each of these motion types is subdivided into four subtypes as specified in Table 1. See Table 2 for details of each of the molecules shown in this figure. (3–4) Comparison of different alignment algorithms using hexokinase (3) and lac repressor (4). (A) standard RMSD fit [6], (B) globally optimized alignment [7] as implemented in SEQUOIA [8], (C) Gaussian-Weighted [11], and (D) sieve-fit [9,10].