Significance
The segmented axial skeleton of vertebrates is composed of an interlinked framework of vertebrae and ribs. During embryogenesis, vertebral precursors known as somites form sequentially from a progenitor tissue known as the presomitic mesoderm to foreshadow the metamerism of the axial skeleton. We have discovered that the cAMP responsive element binding protein (CREB) family of transcription factors operates in combination with Notch and Wnt signaling to instruct the timely scission of presomitic mesoderm into somites with proper anterior/posterior polarities. Thus, the CREB family represents a new and important molecular integrator in axial skeleton development. Our work has potential implications to spinal disorders such as scoliosis.
Keywords: P-CREB, clock and wavefront, somite polarity, A-CREB, CRE-site
Abstract
The segmented body plan of vertebrates is prefigured by reiterated embryonic mesodermal structures called somites. In the mouse embryo, timely somite formation from the presomitic mesoderm (PSM) is controlled by the “segmentation clock,” a molecular oscillator that triggers progressive waves of Notch activity throughout the PSM. Notch clock activity is suppressed in the posterior PSM by FGF signaling until it crosses a determination front at which its net activity is sufficiently high to effect segmentation. Here, Notch and Wnt signaling directs somite anterior/posterior (A/P) polarity specification and boundary formation via regulation of the segmentation effector gene Mesoderm posterior 2. How Notch and Wnt signaling becomes coordinated at this front is incompletely defined. Here we show that the activity of the cAMP responsive element binding protein (CREB) family of transcription factors exhibits Wnt3a-dependent oscillatory behavior near the determination front and is in unison with Notch activity. Inhibition of CREB family in the mesoderm causes defects in somite segmentation and a loss in somite posterior polarity leading to fusions of vertebrae and ribs. Among the CREB family downstream genes, several are known to be regulated by Wnt3a. Of those, we show that the CREB family occupies a conserved binding site in the promoter region of Delta-like 1, encoding a Notch ligand, in the anterior PSM as a mechanism to specify posterior identity of somites. Together, these data support that the CREB family acts at the determination front to modulate Wnt signaling and strengthen Notch signaling as a means to orchestrate cells for somite segmentation and anterior/posterior patterning.
The vertebrate body plan relies on the basic framework of the endoskeleton for support, flexibility, and mobility. To meet these demands, the axial skeleton comprises a series of jointed units of vertebrae, intervertebral discs, and ribs. The metamerism of this organ is laid down as somites, the blueprint for the segmented axial skeleton, during early embryogenesis. In the mouse, somites rhythmically bud off as epithelial spheres from the anterior end of an extending presomitic mesoderm (PSM) approximately every 2 h (1, 2). The newly formed somite is termed somite +1 (S+1) (3). Somites anterior to S+1 are named S+2, S+3, and so on, whereas posterior to S+1 in the PSM are referred to as presumptive somites S0, S−1 and S−2 in descending order (4). The dorsal portion of the somite, the dermomyotome, gives rise to dermis and muscle, whereas the ventral portion, the sclerotome, contains progenitors to the axial skeleton (3). Each sclerotome is polarized such that the anterior half gives rise to the caudal part of the vertebral bodies and distal ribs, whereas the posterior half, the rostral part of the vertebral bodies, vertebral arch, and the rib (5). In the mouse, notochord and sclerotome progenitors form the intervertebral discs which interdigitate the vertebral bodies (6). The anterior/posterior (A/P) polarity of the sclerotome in turn imposes on the segmental organization of the peripheral nervous system (PNS) (7, 8).
Somite segmentation is controlled by a multitiered clock and wavefront program of Notch/FGF/Wnt signaling pathways in the PSM (9, 10). The caudal two thirds of PSM, termed region I, contains posterior-to-anterior FGF and Wnt signaling gradients working in tandem to sustain PSM in an immature state (11–16). The remaining rostral third of PSM, termed region II (from ∼S−2 to S0), is competent for segmental patterning (11, 13, 16, 17). The transition from region I to region II occurs when FGF activity decreases to a lower threshold to define the wavefront (11, 13, 16), where the block in PSM maturation is lifted, creating a determination front. Concomitantly to the FGF wavefront activity, Wnt signaling creates a permissive environment in region I for the segmentation clock to rhythmically operate as traveling waves of gene expression stripes before slowing down in region II to control somite formation upon crossing the threshold set by the determination front (14, 18). Notch signaling is an important output of the clock to synchronize PSM cellular oscillations to counteract the effects of biological noise (e.g., mitosis) (19–21). Wnt3a signaling mediates the expression of the Notch ligand Delta-like 1 (Dll-1) (22), which in turn activates the Notch receptor to form the Notch intracellular domain (NICD) (23). NICD initiates a negative-feedback loop via a transcriptional repressor, Hairy and enhancer of split 7 (Hes7) and a modifier, Lunatic fringe (Lfng) in the initial cycle to create periodic waves of NICD throughout region I to temporally synchronize PSM cells (23–25). Importantly, in the mouse, Notch signaling is also essential for translating the clock message into somite A/P polarity and boundaries in region II (26, 27). The intermediate NICD stripe (∼S−2/S−1) in combination with Wnt signaling near the determination front is critical (26). Here, Notch activity reaches a high level of net output to overcome the repressive effects of the wavefront and synergizes with the Wnt-regulated T-box 6 (Tbx6) transcription factor to activate an expression stripe of the segmentation effector gene, Mesoderm posterior 2 (Mesp2) (26–28). Subsequently, Mesp2 forms a graded band originating from the anterior compartment of a nascent somite followed by its inactivation by Ripply2, leading to somite boundary formation and A/P specification (29, 30). How Notch/Wnt signaling is coordinated in region II to instruct A/P patterning and boundary formation via Mesp2 is not completely understood.
We discovered that mice mutant for the transcription factor cAMP responsive element binding protein (CREB) sometimes displayed somite segmentation defects that could be attributed to dysregulated Notch signaling. CREB is the founding member of the CREB family, which also includes activating transcriptional factor 1 (ATF-1) and cAMP responsive element modulator (CREM) (31–34). Together they constitute a distinct subfamily that belongs to a large family of basic leucine zipper transcription factors (35). The three CREB family members form homo- or heterodimers with each other via their leucine zippers, whereas their basic domains bind to a full cAMP responsive element (CRE) site (TGACGTCA) or a half CRE-site (TGACG) (31, 36–41). The transcriptional activity of this family depends on stimulus-induced phosphorylation of CREB, CREM, and ATF-1 at Ser133 (P-CREB), Ser117 (P-CREM), and Ser63 (P-ATF-1) in their kinase inducible domain (32, 42, 43). Phosphorylated kinase inducible domain associates with coactivators for recruitment of transcriptional machinery to activate downstream genes (44, 45). Here we describe an essential role for the CREB family of transcription factors in somite segmentation by using a dominant-negative approach to reduce the combined activity of the CREB family in the mesoderm. We propose that this family orchestrates somite boundary formation and posterior polarity by modulating Wnt signaling and increasing the net Notch activity at the determination front.
Results
Dynamic CREB Family Activity Is Localized Near Determination Front in Region II of PSM.
During our previous investigation of CREB in myogenic gene induction (46), we observed a rare population of CREB mutants with abnormally shaped somites (Fig. S1 B and D). By whole-mount skeletal preparation, we also found that mutant neonates exhibited sporadic vertebral fusions (Fig. S1 G and I). These data implied a role for CREB in somite segmentation and inspired us to reexamine CREB activity during somitogenesis. To monitor the activity of the CREB family, we used whole-mount immunostaining using phospho-CREB (P-CREB)-specific antibodies (Fig. S2 A–D and F), which have served as a reliable proxy for CREB activity (46–48). Consistent with previous reports (46, 49), CREB protein was ubiquitously distributed in embryos, including somites and PSM at embryonic day (E) 10.5 as an example (Fig. S2E), whereas P-CREB was present between E8.5 to E12.5 as one or two distinct bands in region II and in the dermomyotome (Fig. S2 B–D and F). No detectable P-CREB stripe was observed in the PSM at E7.5 (Fig. S2A).
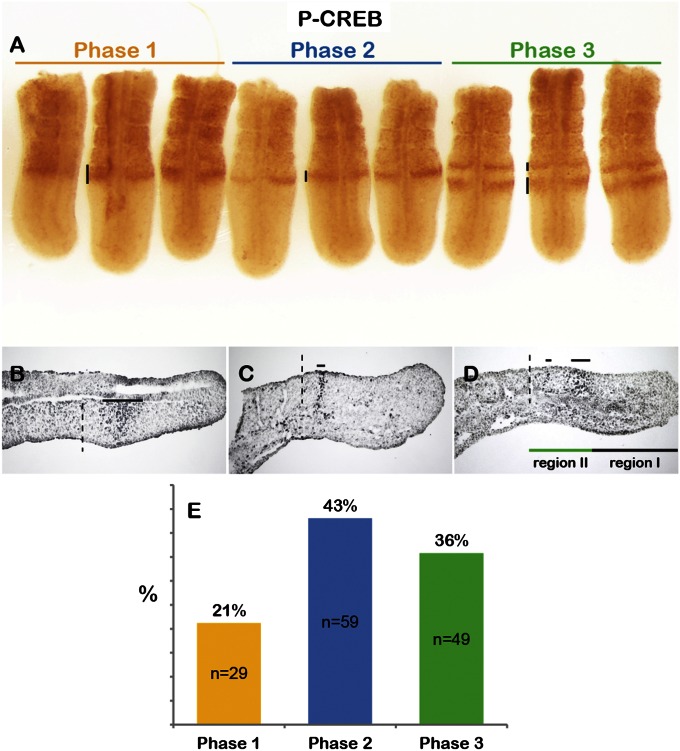
Three distinguishable P-CREB patterns emerged in region II of the PSM, which we named Phases 1 to 3 (Fig. 1 A–D). In Phase 1, P-CREB showed a broad band near the S−1 and S0 interface (n = 29/137; Fig. 1 A, B, and E). In Phase 2, only a narrow stripe in the posterior part of S0 was found (n = 59/137; Fig. 1 A, C, and E). In Phase 3, the stripe in the posterior part of S0 was further narrowed and an additional broad band appeared near S−2 and S−1 (n = 49/137; Fig. 1 A, D, and E). P-CREB stripes were not detected in region I and tailbud except for a few scattered cells, even when overdeveloped in the chromogenic substrate. The dynamic stripes likely represent combined activity of all three CREB family members, as they are expressed during the stages examined (46, 49) and P-CREB antibodies are predicted to recognize all three. The PSM P-CREB patterns were missed in our previous study likely caused by their dynamic changes and the use of transverse sections for assay (46). From here on, we refer to the P-CREB staining patterns as the CREB family activity.
Fig. 1.
Dynamic CREB family activity is localized to region II of the mouse PSM. (A) Whole-mount P-CREB staining on E10.5 mouse tails (dorsal view, anterior to the top). P-CREB signal in the PSM is indicated by black lines: Phase 1 (orange), Phase 2 (blue), and Phase 3 (green). (B–D) Sagittal sections of tails stained for P-CREB; dotted lines, S0 and S+1 interface; solid lines, P-CREB signal: (B) Phase 1, (C) Phase 2, and (D) Phase 3, below tail section. Black line depicts region I; green line is region II. (E) Graph summarizes the percentage (y axis) of each Phase (x axis) from 137 samples; n of each Phase indicated in bars.
CREB Family Activity Cycles in Region II of PSM.
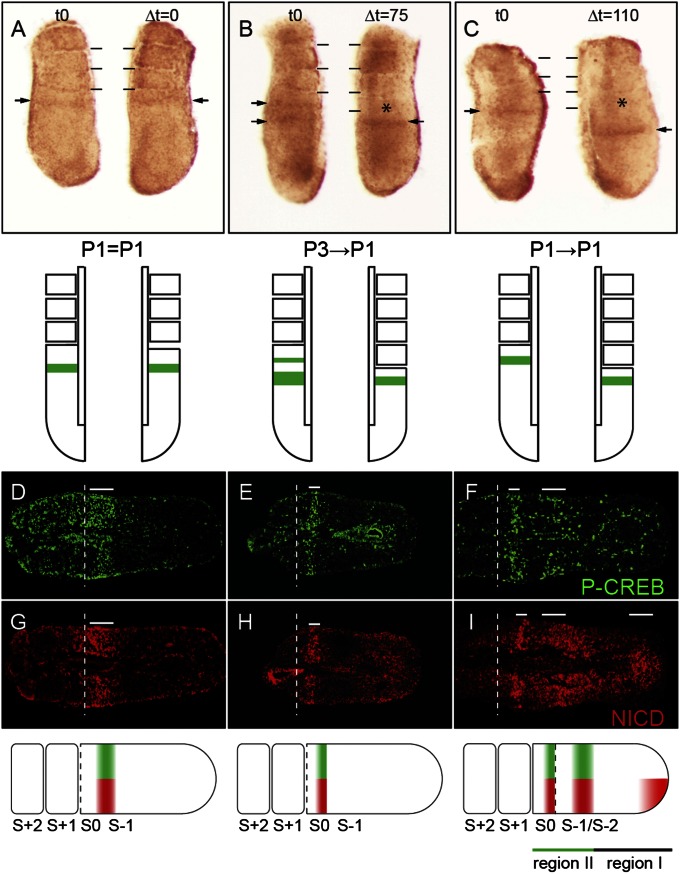
To test whether the dynamic CREB family activity in the PSM represents oscillation, we bisected E10.5 embryonic tails (n = 30) longitudinally, fixing one half to capture the state of the activity at t = 0 and culturing the contralateral half for various lapsed time points (∆t). As a control, when both halves were fixed simultaneously (∆t = 0), no difference was observed as both halves displayed the same CREB family activity state (Fig. 2A). When the isolated PSM was at Phase 3, we captured a transition to Phase 1 in the experimental half cultured for 75 min (Δt = 75) along with a newly formed somite (Fig. 2B), supporting oscillating CREB family activity. To observe one complete revolution of activity during one round of somitogenesis, both halves were cultured until a somite was formed. One half was fixed and the other half was further cultured until a second somite was generated (Δt = 110). Both halves were found to be at Phase 1 (Fig. 2C). Thus, a new round of somite formation appears to begin at Phase 1 and one cycle coincides with a single somite budding event, revealing segmentation clock-like characteristics.
Fig. 2.
CREB family activity cycles with the Notch clock in the PSM. (A–C) P-CREB staining of E10.5 PSM halves. (A) Left and right halves fixed immediately (t0) are both in Phase 1 (P1). (B) Phase 3 (t0, Left) to Phase 1 (Δt = 75, Right) transition (P3→P1) is found after culture for 75 min. (C) Phase 1 P-CREB pattern (t0, Left) completes one full cycle (P1→P1) in the right half cultured for 110 min (Δt = 110). Diagrams for results in A–C are directly below; solid lines indicate somite boundaries; asterisk indicates new somite; arrows indicate P-CREB stripes. (D–I) Immunofluorescence of P-CREB (green; D–F) and NICD (red; G–I) on E10.5 tail horizontal sections (anterior to the left): (D and G) Phase 1 (n = 3/20), (E and H) Phase 2 (n = 10/20), and (F and I) Phase 3 (n = 7/20) are paired adjacent sections; white dotted lines indicate the boundary between S0 and S+1; white solid lines indicate areas of P-CREB and NICD signal. Schematics depicting that P-CREB (green) oscillates in parallel with NICD (red) are directly below. In the schematic illustration (Bottom), black line depicts region I and green line is region II.
To determine how the CREB family activity is related to the Notch clock (23), we performed comparative immunofluorescence on E10.5 PSM stained for P-CREB and NICD. Examination of the three phases of CREB family activity relative to NICD on immediate adjacent sections revealed that they are in sync in region II (Fig. 2 D–I). Despite the shared cycling relationships in region II of the PSM, very few positive cells for CREB family activity were found to overlap with NICD (Fig. 2 D–I) in region I. Importantly, in Phase 3, the broad P-CREB stripe coincides with the NICD stripe near the determination front (S−2/S−1; Fig. 2 F and I). These data indicate that CREB family activity stripes in region II are potentially linked to the Notch clock.
CREB Family Directs Somite Boundary Formation and Maintains Somite Epithelium.
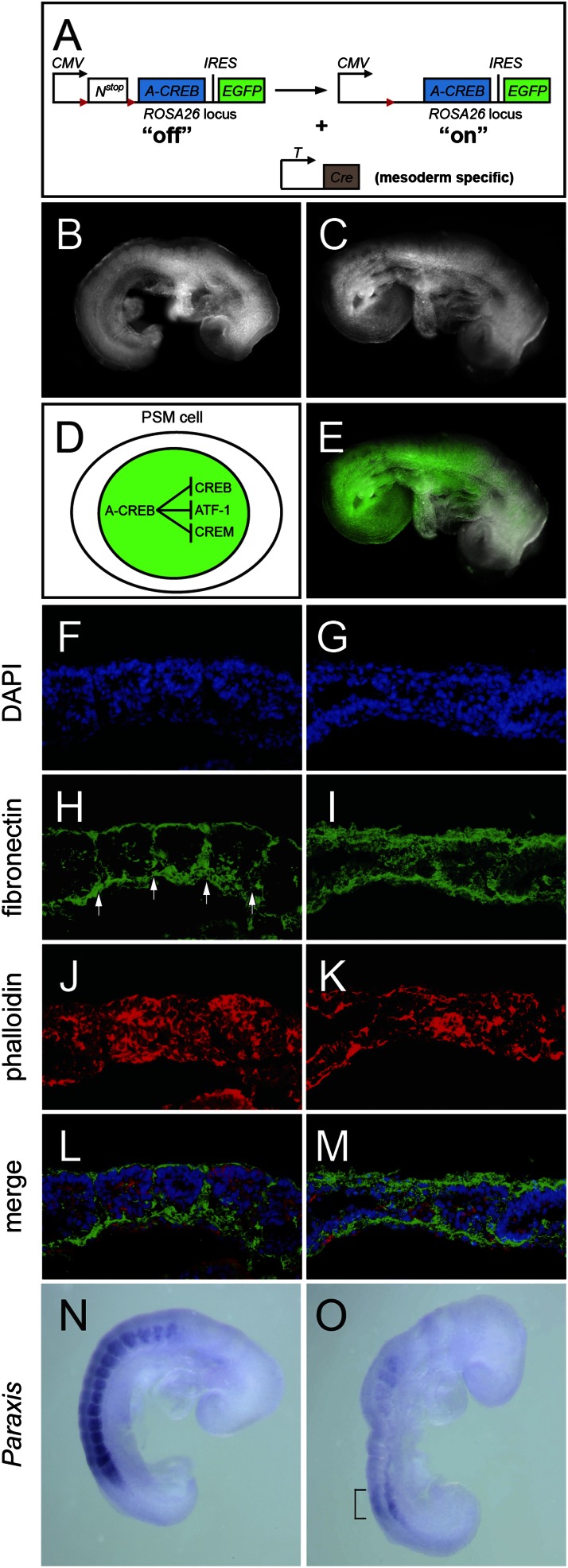
The patterns of CREB family activity suggested that they play a role in somite segmentation. Testing CREB function in this process by germ-line mutants had been a challenge as the segmentation phenotype showed very low penetrance (Fig. S1E), likely as a result of redundancy with other family members. We generated CREB−/−;CREM−/− mutants but failed to increase the frequency of mutants with segmentation defects. Conversely, CREB−/−;ATF-1−/− mutants die before implantation and CREB−/−;ATF-1+/− mutants are developmentally delayed and die at approximately E9.5 (49). To circumvent this, we designed a tissue-specific dominant-negative approach. A widely used and highly selective pan–dominant-negative reagent to the CREB family is acidic-CREB (A-CREB), which heterodimerizes with the leucine zipper motif, whereas the acidic patch traps the positively charged basic residues of the CREB family to prevent DNA binding (50, 51). In vitro studies have shown the specificity of this reagent: 1 M equivalent of A-CREB effectively interrupts CREB binding to a CRE-site, whereas 100 M excess of A-CREB cannot abolish DNA binding of other basic leucine zipper subfamilies (35). We generated a conditional A-CREB-IRES-EGFP allele at the ROSA26 locus (52), referred to as ROSAAC (Fig. 3A and Fig. S3). The CMV/β-actin enhancer assures high levels of expression in the mesoderm upon the removal of a loxP-flanked Neo-stop cassette by the T-Cre driver (53) (Fig. 3 A and D and Fig. S3). Approximately 40% of the ROSAAC/+;T-Cre adults have fusions of the cervical vertebrae (Fig. S3E), suggesting that one copy of the ROSAAC was insufficient to inhibit all CREB family member activity. To increase the dosage, we generated ROSAAC/AC;T-Cre embryos (referred to as mutants later). We observed robust live EGFP signal reflecting A-CREB expression in the PSM and somites compared with controls (Fig. 3 B and E). Pleasingly, we found that mutant somites had poorly defined boundaries along the body axis (Fig. 3C), supporting that the CREB family acts redundantly to instruct somite segmentation.
Fig. 3.
The CREB family is essential for somite patterning. (A) Diagram depicting conditional expression of A-CREB and EGFP driven by T-Cre after removing the floxed Neo-stop (Nstop) cassette in ROSAAC. (B and C) Bright-field images of E8.75 ROSAAC/AC (control) and ROSAAC/AC;T-Cre (mutant) embryos, respectively. (D) Schematic of nuclear EGFP marking of A-CREB–expressing cells. (E) Mutant embryo image (C) merged with EGFP fluorescence. (F–M) Comparison between somite morphologies of control (F, H, J, and L) and mutant (G, I, K, and M) embryos; sagittal sections, anterior to the left: (F and G) DAPI-stained nuclei; (H and I) fibronectin-stained somite boundaries (white arrows); and (J and K) phalloidin-stained actin. (L) Merged image from F, H, and J. (M) Merged image from G, I, and K. (N and O) Paraxis expression is reduced in mutants (O) compared with controls (N), and bracket indicates residual expression in the mutant.
To determine whether the morphological defects are associated with abnormalities in somite boundaries and epithelial structures, we examined the organization of the ECM, nuclei, and actin of mutant somites. Contrary to controls (Fig. 3 F and J), E8.5 mutant somites (n = 4) lost their compact epithelial morphology, as the nuclei and actin-labeled cells displayed a scattered appearance (Fig. 3 G and K). This was corroborated by misaligned fibronectin organization between adjacent somites (Fig. 3I). Histological analysis of E9.5 mutants (n = 3) revealed that mutant somites were smaller in size and lacked the well separated epithelia seen in controls (n = 3; Fig. S4 A–D). This defect is strikingly reminiscent to the rare CREB severe mutant (Fig. S1 B and D). Furthermore, mutants displayed reduced myotomal mass (Fig. S4D), consistent with a role for CREB in myogenesis (46). We next examined whether the CREB family regulates the somite epithelial determinant gene Paraxis (54). Control and mutant embryos displayed characteristic Paraxis expression in region II (Fig. 3 N and O, bracket). However, Paraxis became significantly reduced in mutant somites compared with controls (Fig. 3 N and O). Thus, the failure of maintaining Paraxis expression helps to explain the poor somite epithelial organization observed in the mutant.
Mutants Compromised for CREB Family Activity Display Extreme Skeletal Dysplasia.
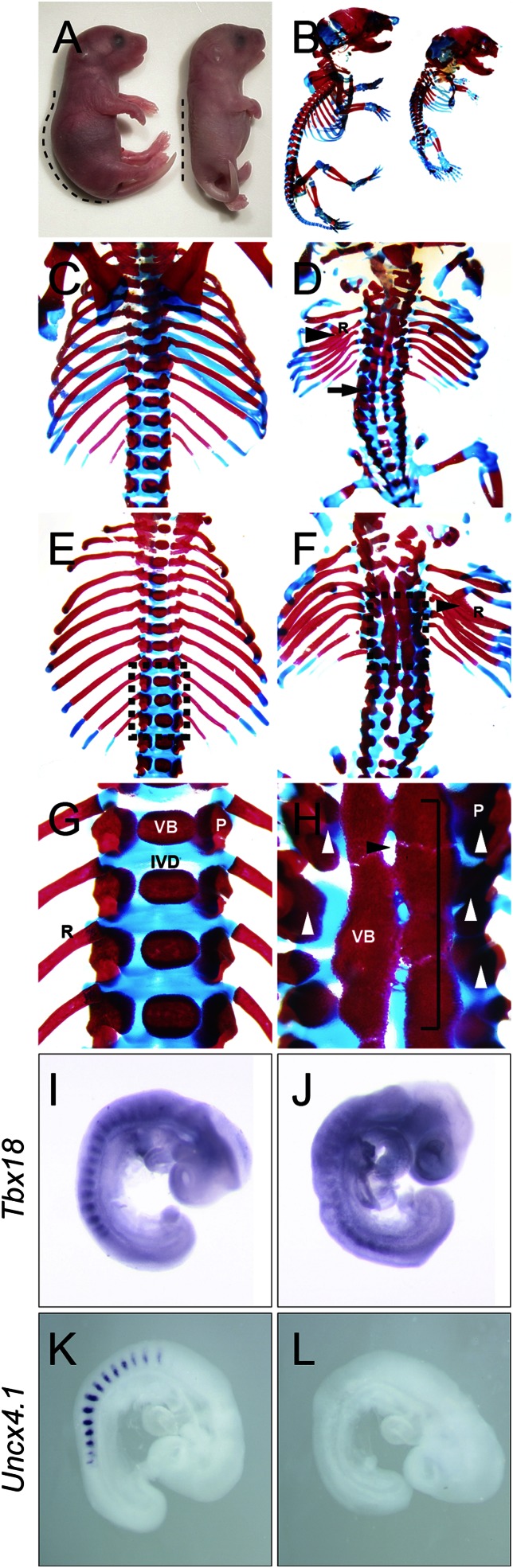
All mutant animals died at birth, likely of respiratory failure. Mutants were identifiable from control littermates by their straightened postures and kinked tails (Fig. 4A). The axial skeleton of mutant neonates was conspicuously defective along the cervical, trunk, and lumbar regions, and, to a lesser extent, in the tail (Fig. 4B). Closer examination of the mutant vertebral column (Fig. 4 D, F, and H) revealed fusions of the neural arches and vertebral bodies and a loss of intervertebral discs, compared with controls (Fig. 4 C, E and G). The lack of intervertebral discs is likely a result from defects in somite polarity (17) rather than a loss of the notochord, as the expression of a key notochordal signaling molecule, Shh (6, 55), was unchanged in the mutant (Fig. S6 A and B). The sclerotome has distinct A/P halves that contribute to the metamerism of the axial skeleton and PNS (5–8). At E11.5, mutant embryos (n = 4) displayed long stretches of mesenchymal patches that did not segregate into repeated intervals (Fig. S4 F and H), whereas control embryos (n = 5) contained dense metameric condensations along the A/P axis (Fig. S4 E and G). Consequently, the mutant dorsal root ganglia were fused to each other (Fig. S4 I and J) and axon bundles were not properly spaced compared with control embryos (Fig. S4 K and L). This was further confirmed by whole-mount neurofilament immunostaining whereby mutant axons were frayed throughout the somite (n = 3; compare Fig. S5 A and B). These results demonstrate that reducing CREB family function in the mesoderm impedes the segmental organization of the somite, resulting in the fusion of the axial skeleton and disorganization of the associated sensory neurons and axons.
Fig. 4.
Skeletal defects in mice deficient for CREB family activity during somitogenesis. (A) ROSAAC/AC (control, Left) and ROSAAC/AC;T-Cre (mutant, Right) neonates (P0); dotted lines mark the postures. (B–H) Skeletal preparations stained with Alizarin red and Alcian blue. (B) Entire skeletons of control (Left) and mutant (Right). Control (C, E, and G) and mutant (D, F, and H) axial skeletons: (C and D) dorsal view; (E and F) ventral view; and (G and H) magnified views of the rectangular area in E and F, respectively. Arrowhead indicate bifurcation of rib (marked as “R”), and arrow indicate fusion of the lamina. White arrowheads indicate fusions of pedicles (marked as “P”), and the bracket indicates fusions of vertebral bodies (VB). Control in G has intervertebral discs (IVD), and the black arrowhead in the mutant indicates a loss of intervertebral discs (H). (I–L) WISH analysis of Tbx18 (I and J) and Uncx4.1 (K and L) in controls (I and K) and mutants (J and L).
To determine the underlying molecular mechanism, we examined the expression of Tbx18 and Uncx4.1, genes that sustain the anterior and posterior somite identities, respectively (56, 57). Indeed, posterior somite specification is defective in mutants as Uncx4.1 transcripts were lost in the PSM and somites (Fig. 4L) to an expansion of Tbx18 expression, albeit also reduced (Fig. 4J). Thus, the axial skeletal defects of mutants can be explained by the dysregulation of key somite polarity genes.
CREB Family Activates Defined Transcriptome in PSM.
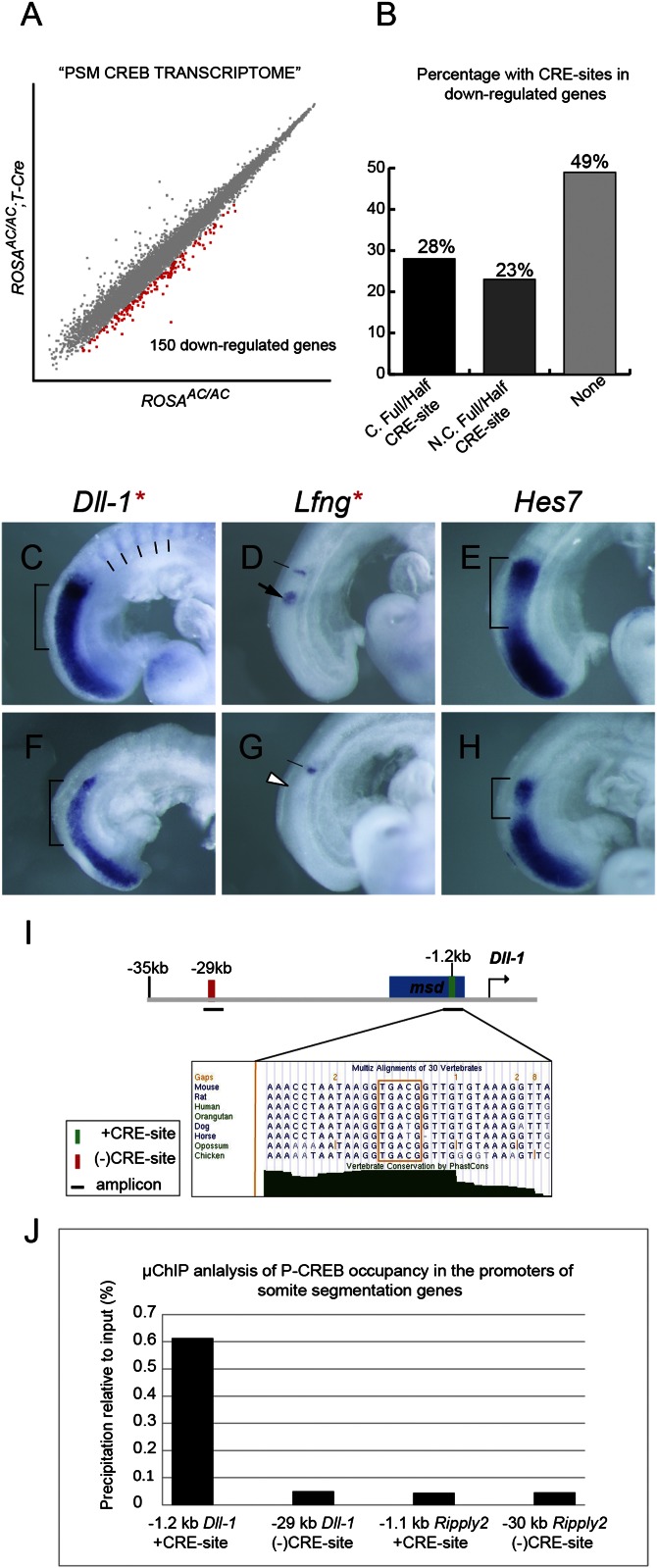
The morphological and molecular defects must be a consequence of compromised CREB family transcriptional activity in the anterior PSM. We therefore decided to uncover its downstream target genes. By microarray analysis of pooled samples of region II from mutants and controls, we identified 328 genes that were differentially expressed (Fig. 5A). We refer to this group of genes as the PSM CREB transcriptome, which contains genes in various functional categories. Of this transcriptome, 150 genes were down-regulated more than 1.5 fold in mutants and ∼28% of them contain putative full or half conserved CRE-sites in their promoters among human, mouse, and rat genomes (58) (Fig. 5B and Table S1). Many of these CRE-sites identified were within a few thousand bases upstream of transcription start sites (TSSs), implicating them as primary CREB family targets. Although we collected stage-matched embryos for microarray analysis, the pooled region II PSMs were likely at different phases of P-CREB pattern. Hence some downstream genes might not have been uncovered. Importantly, by comparing our PSM CREB transcriptome to the existing segmentation clock network identified by Dequent et al. (59), we found several key segmentation genes associated with Notch and Wnt signaling pathways (Table S1).
Fig. 5.
The CREB family controls somite morphogenesis through Dll-1/Notch signaling. (A) Scatter plot of log2-transformed hybridization signal intensity values between ROSAAC/AC;T-Cre (y axis) mutants, and ROSAAC/AC (x axis) controls. A total of 150 genes (red squares) were found to be down-regulated. (B) Bar graph tabulates percentage of down-regulated genes with conserved (marked as “C”) full/half CRE-sites, nonconserved (N.C.) full/half CRE-sites, and none. (C–H) WISH analysis of Dll-1 (C and F), Lfng (D and G), and Hes7 (E and H) in controls (C–E) and mutants (F–H): brackets indicate altered Dll-1 levels and Hes7 domains between controls and mutants; lines in control (C) marks Dll-1 expression in posterior somite halves; white arrowhead in mutant (G) indicates strongly reduced Lfng cyclic expression; red asterisks indicate genes with a putative CRE-site. (I) Diagram of the Dll-1 promoter and −35 kb upstream of the transcriptional start site. Blue box is the msd enhancer. Magnified box is a multiple sequence alignment of a conserved +CRE-site (orange box); green bar indicates CRE-site; red bar indicates (−)CRE-site; solid lines indicate amplicons. (J) Bar graph of μChIP quantification. P-CREB occupation at the −1.2 kb CRE-site (0.614%) in the Dll-1 promoter is 12.3-fold higher than the (−)CRE-site control (0.050%; −29 kb). P-CREB binding to the −1.1 kb CRE-site (0.044%) in the Ripply2 promoter is comparable to the (−)CRE-site control (0.046%; −30 kb).
CREB Family Controls Notch Signaling by Directly Regulating Its Ligand Dll-1.
Mixed P-CREB phases of embryos along with the low dynamic range of detection by microarray prompted us to validate candidate downstream genes by whole-mount in situ hybridization (WISH) for a better visual of their spatial and temporal changes. We first focused on the Notch pathway genes: Dll-1 and Lfng. We verified that the Notch ligand Dll-1 was down-regulated with a “salt and pepper”-like pattern in region II of the mutant PSM and lost in the posterior half of mutant somites, compared with controls (Fig. 5 C and F). The extent of reduction by WISH appears to be more than that obtained by microarray (−1.7 fold). Cyclic expression of Lfng, a Notch-induced readout of the segmentation clock (23, 25), was also disrupted, displaying reduced levels with a salt and pepper pattern near the transition of region I/II (Fig. 5 D and G and Fig. S6 C–H, white arrowheads). Its expression at the most anterior band is similarly affected (Fig. 5 D and G and Fig. S6 C–H, black lines). Its reduction appears greater than that obtained by microarray (−1.65 fold). We did note that Lfng was initiated in the caudal of region I in Phase 3 of the Notch clock cycle (Fig. S6 C and F). To extend this observation, we examined another Notch-induced cycling gene, Hes7 (24). Hes7 expression pattern in region I was unchanged, but the length of its oscillatory field in region II was shrunken (Fig. 5 E and H). The altered expression of Lfng and Hes7 likely resulted from reduced Notch signaling. Thus, CREB family activity impacts Notch signaling by maintaining critical expression levels and spatial patterns of its positive (i.e., Dll1) and negative (i.e., Hes7/Lfng) regulators in region II.
To assess whether a given gene in the PSM CREB transcriptome is directly regulated by the CREB family, we used micro-ChIP (µChIP) to determine whether P-CREB binds to its promoter in vivo. We selected genes for testing based on three criteria: First, a CRE-site(s) within 2,000 kb of the promoter; second, clearly reduced gene expression confirmed by WISH; and third, a documented role similar to the CREB family in somitogenesis (60, 61). Dll-1 fits all three criteria, especially considering that the CREB family and Dll-1 mutants have the same defects in A/P polarity and somite segmentation (60, 61). For chromatin binding, we tested the conserved CRE-site located −1.2 kb from the TSS, which is positioned in a mesoderm enhancer element (msd; Fig. 5 I and J) shown to recapitulate Dll-1 expression in the PSM (62). By using the percent input method, we quantified in vivo occupancy of the region containing predicted CRE-site vs. a reference region located −29 kb without a CRE-site, (−)CRE. Indeed, P-CREB binding to the CRE-site in the Dll-1 msd was ∼12.3 fold higher than that of the (−)CRE region (Fig. 5J). This data strongly supports that the CREB family directly regulates Dll-1, thereby increasing net Notch signaling in region II of the PSM.
CREB Family Coordinates Position of Wnt Signaling Gradient in Region II and Segmentation Machinery in PSM.
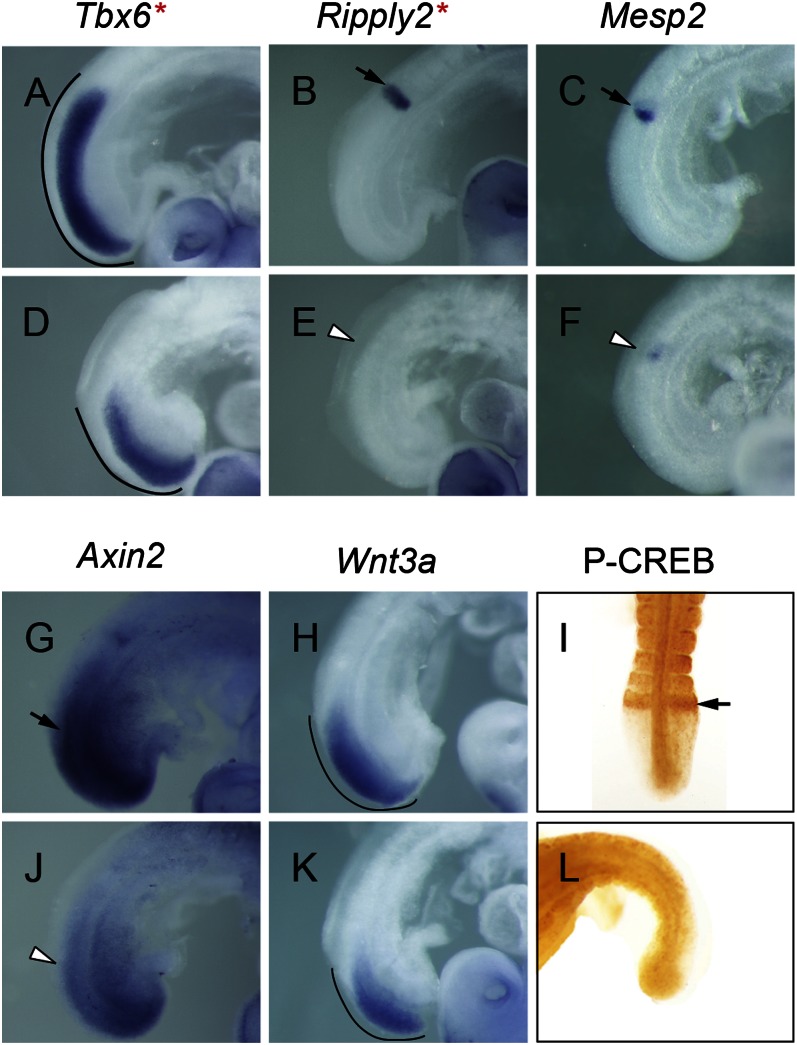
The down-regulation of Dll-1, a Wnt-regulated gene (22), encouraged us to examine this signaling pathway further. As mentioned earlier, segmentation genes controlled by Wnt signaling are also noted: Tbx6 and Ripply2 (Table S1). WISH analysis confirmed that Tbx6 expression was severely down-regulated in region II of the mutant PSM compared with the controls (Fig. 6 A and D). The tight band of Ripply2 expression in the control was virtually undetectable in the mutant (Fig. 6 B and E). The extent of down-regulation of Tbx6 and Ripply2 assessed by WISH appears more reduced than that by microarray (−1.75 and −3.0 fold, respectively), and supports that Wnt signaling in region II is defective in mutants.
Fig. 6.
The CREB family modulates a segmentation network in region II. (A–H, J, and K) WISH analysis. Tbx6 (A and D), Ripply2 (B and E), Mesp2 (C and F), Axin2 (G and J), and Wnt3a (H and K) in controls (A–C, G, and H) and mutants (D–F, J, and K). Note that reduced Mesp2 expression persists as a stripe in mutant compared with control. Curved lines mark the length of the graded PSM expression pattern of genes; black arrow indicates control expression pattern; white arrowhead indicates altered expression pattern in mutant; red asterisks indicate genes containing putative CRE-sites. (I and L) P-CREB staining. Arrow indicates the P-CREB stripe in Wnt3avt/+ control (I), which is missing in the Wnt3avt/vt mutant (L).
By the same criteria for choosing to test Dll-1 as a direct target of the CREB family, we chose Ripply2 (29, 30, 63) for μChIP analysis. We tested the occupancy of P-CREB on the conserved CRE-site located −1.1 kb from the TSS of Ripply2 vs. a region without a CRE-site at −30 kb. Curiously, we did not detect a significant occupancy at the −1.1 kb CRE-site (Fig. 5J). We presume that the CREB family uses a different or cryptic CRE-site to regulate Ripply2 or that the CREB family indirectly regulates Ripply2.
The reduction of the Wnt-regulated gene Tbx6 in region II (Fig. 6D) prompted us to examine whether the Wnt3a signaling range was affected. We first examined whether a direct downstream cyclic gene of Wnt3a, Axin2 (12), was misregulated. Relative to the control, Axin2 oscillation was down-regulated throughout region II in mutant PSM except for the most posterior area of region I (Fig. 6 G and J). The Wnt3a expression domain was maintained but slightly reduced in the mutant PSM relative to the control (Fig. 6 H and K). Thus, the Wnt3a signaling level appears decreased near the determination front in mutants, suggesting that CREB family activity acts downstream of Wnt3a reception to mediate signaling. Indeed, the striped P-CREB patterns were no longer observed in the PSM of the Wnt3avt/vt embryo (Fig. 6 I and L), in which Wnt3a activity is much reduced (12). Together these data suggest that CREB family activity depends on Wnt3a signaling, which then feeds into a regulatory pathway to maintain Wnt3a downstream genes in region II, possibly as a way to refine the range of the Wnt signaling gradient.
Given that Mesp2 is downstream of Tbx6 and Notch signaling (27, 64) and upstream of Ripply2 (29), we examined its expression. Although not found to be significantly reduced by microarray, Mesp2 is in fact diminished but not lost in the mutant. Although Mesp2 expression stripes cycles in the anterior halves of S−1/S0 in control embryos (Fig. 6C and Fig. S6 I and J), we noted that all mutant embryos displayed identically weak and loosely organized Mesp2 banding pattern (n = 5; Fig. 6F) indicative of persistent rather than dynamic expression. Thus, A/P polarity and boundary defects in mutants can be interpreted by reduced net Notch signaling activity and Wnt signaling range, leading to misexpression of Mesp2.
Discussion
We demonstrate here that CREB family activity matches Notch clock oscillations in region II, but not in region I (Fig. 2 D–I). We also provide data implicating that Wnt3a signaling involves phosphorylation of the CREB family in region II. Here, the CREB family likely mediates Wnt3a signaling by increasing the expression of its downstream genes Axin2, Dll-1, and Tbx6. The canonical Wnt/β-catenin pathway is also essential for somite segmentation (12, 14, 18). A posterior-to-anterior nuclear β-catenin gradient in region I ends near the determination front to maintain PSM in an immature state and defines the oscillatory field for the segmentation clock (12, 14, 18). Thus, the CREB family activity represents a new branch of Wnt3a signaling to confer PSM maturation at the determination front. How the CREB family and β-catenin coordinate to effect segmentation awaits future studies. Wnt3a has also been shown to effect phosphorylation of the CREB family for myogenic gene expression in the dermomyotome (46), revealing a sequential switch from Wnt-CREB signaling in the PSM along the anteroposterior axis to signaling in the somite along the dorsoventral axis. Interestingly, in cell culture studies, induction of CREB phosphorylation has been documented to peak within 30 min, followed by a burst in gene expression within a 2-h window (65), similar to the cycling time observed in the PSM. We therefore propose that P-CREB results from the anterior end of the Wnt3a signaling gradient and turns over as the cells move away from its influence. Because Wnt3a is abundant in the posterior PSM, the lack of CREB family activity there indicates a suppressive cue in region I. CREB family activity is also preferentially suppressed in the anterior but maintained in the posterior somite S0. One possibility is that it also responds to spatial cues emanating from the somites such as retinoic acid signaling (66, 67), which is thought to counteract the wavefront to fine tune the determination front.
We determined the function of the CREB family in somitogenesis by devising a mesoderm-specific conditional approach to inhibit all three members via a pan–dominant-negative transgene, A-CREB, in combination with the T-Cre driver. Curiously, we did not observe gastrulation defects described for ATF-1;CREB mutants (49). We presume that, after T-Cre–mediated recombination, A-CREB expression accumulates to an effective inhibitory level only well after cells have already left the primitive streak. Dosage-dependent defects of this family were evident from their compound mutants (49, 68). We also found more severe skeletal defects by doubling A-CREB dosage, further extending its utility. We therefore envision that the ROSAAC allele is a powerful modular tool in combination with a vast number of tissue-specific and inducible Cre-driver lines to determine CREB family function in other contexts.
Comparative expression profiling between ROSAAC/AC and ROSAAC/AC;T-Cre region II PSMs led us to discover the PSM CREB transcriptome. Extensive genomewide analysis revealed that CREB can occupy in the range of 4,000 to 6,000 mammalian CRE-sites (58, 69). This large number of CRE-sites gives CREB an enormous flexibility in activating diverse genetic programs. In a given cellular or tissue context, not all CRE-sites are accessible to the CREB family. For example, Ripply2 is strongly down-regulated in ROSAAC/AC;T-Cre embryos, but P-CREB binding to a putative CRE-site within its promoter is not detected. Conversely, we did find that Dll-1 is down-regulated in mutant embryos and that P-CREB binds to a conserved CRE-site in the msd enhancer of Dll-1 (62), demonstrating that the CREB family directly contributes to Dll-1 expression levels in region II. The CREB family likely cooperates with Tbx6 and LEF/TCF at the Dll-1 promoter (22). Reduced Dll-1 in mutants presumably fails to activate a net Notch activity required to drive segmentation and polarity specification as evidenced by weakened Mesp2 and Lfng expression. In addition, Dll-1 is also known to maintain somite epithelium by sustaining high levels of Paraxis expression after somite segmentation (60). Consistently, ROSAAC/AC;T-Cre embryos have epithelialization defects accompanied by strongly reduced Paraxis and Dll-1 expression in the somite. The reduction of Paraxis expression appears more severe in the ROSAAC/AC;T-Cre embryos (Fig. 3O) than in the Dll-1 mutant embryos (60), suggesting that the CREB family controls Paraxis expression only partially through Dll-1. Because CREB family and Paraxis mutants have the same epithelial defect but opposite defects in A/P polarity patterning (54, 70), Paraxis misregulation can only explain the former in the CREB family mutant. The precise mechanism of how the CREB family controls Paraxis in the mature somite requires further investigation. On the contrary, the similarities between Dll-1 and CREB family mutants supports that CREB family directly controls Dll-1 mediated processes in the PSM and partially in the somites.
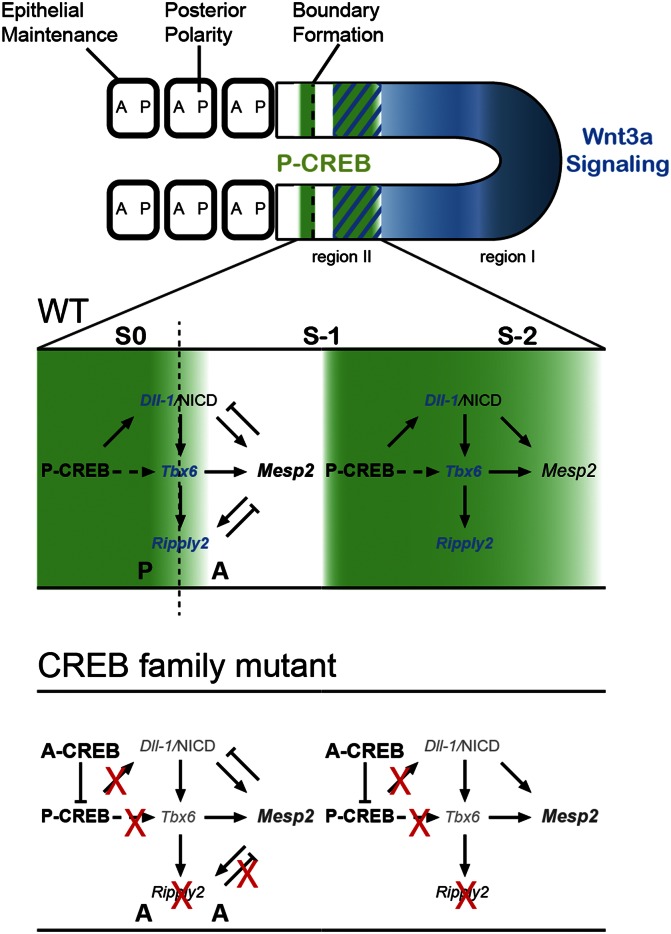
One of the most intriguing results we found was the loss of somite boundaries and posterior patterning upon the removal of CREB family activity from the PSM. Given that the CREB family acts directly upstream of Dll-1, we deduced that this family may be critical for Notch clock activity, which is essential for A/P patterning and somite segmentation (23, 26). This is reflected by the narrowed Hes7 field, interrupted Lfng cycling, and persistent Mesp2 expression in region II of the mutant. The current view of the A/P patterning and segmentation machinery has linked Notch activity together with Tbx6 at the determination front to trigger Mesp2 expression first as a stripe that spans the length of a somite (26, 27). This is followed by Mesp2 and Tbx6 synergistically activating Ripply2 to restrict the Mesp2 stripe to the anterior of S−1 and finally to shut it off before segmentation in S0 (14, 29). The reduced yet sustained Mesp2 pattern in the ROSAAC/AC;T-Cre embryos can be explained by the concomitant reduction of Dll-1 and Tbx6. The loss of Ripply2 expression in the mutant is likely a consequence of reduced Tbx6 and/or Mesp2, thus causing the expansion of the anterior compartment. This is further supported by the loss of Uncx4.1 expression and expanded Tbx18 expression. It follows that the somite boundary and polarity defects reported for Dll-1 and Ripply2 single mutants (29, 60, 61) resemble those of the ROSAAC/AC;T-Cre embryos. Thus, the CREB family acts upstream of Dll-1 in region II to contribute to the activation of Notch and Tbx6 to effect cyclic Mesp2 and Ripply2 activities for generating somite boundaries and establishing polarity. In light of our data, we propose that the spatial–temporal activity of the CREB family serves as an integration point to modulate the Wnt3a signaling gradient and increase net Notch signaling as a means to translate the activities of the segmentation clock and wavefront in cells to undergo segmentation (Fig. 7).
Fig. 7.
The CREB family forms a core segmentation module in somitogenesis. In our model depicted at Phase 3, CREB family activity (P-CREB) oscillation (in green) has critical roles in somitogenesis: to mediate Wnt3a signaling at the anterior end of its gradient (blue) in the P-CREB domain (green/blue hatched marks), coordinate the segmentation machinery in region II, and maintain somite epithelium. In WT (magnified, Middle), P-CREB (green area) directly regulates the expression of Dll-1 at S−2/S−1 and mediates the expression of Tbx6. In turn, Dll-1/Notch signaling and Tbx6 synergize to activate Mesp2 to define the anterior somite half (white band) of S−1. Tbx6 and Mesp2 subsequently activate Ripply2 to refine Mesp2 expression. The coordination of P-CREB and Mesp2 in region II drives the formation of somite boundaries and A/P patterning at S-1 and S0. Black dotted line marks the future somite boundary. (Bottom) In the CREB family mutant, there are reduced Dll-1/Notch signaling and Tbx6 expression, sustained albeit at a lower level of Mesp2 stripe, and loss of Ripply2, leading to anteriorized somites. Blue text represents Wnt-regulated genes. Light text represents reduced expression. Symbols in regulatory loop: arrow for direct activation, dotted arrow for indirect or unknown activation, T for inhibition, and X for lost regulatory pathway or gene expression. A, anterior somite half; P, posterior somite half.
Although we establish a basic framework to interpret how the CREB family activity fits into the segmentation network, additional genes in the PSM CREB transcriptome likely contribute to the mutant phenotypes. Future investigations will provide new threads toward building a comprehensive regulatory circuitry. Disruption of this circuitry leads to compromised Notch signaling, which has a direct implication to human spine disorders. Mutations in MESP2, DLL3, or LFNG have all been associated with vertebral malformations such as spondylocostal dysostosis (71–73). As a result of the redundancy between CREB family members, mutations in each gene alone may not cause vertebral defects. On the contrary, because the CREB family activity can be influenced by many stimuli, we imagine that idiopathic sporadic scoliosis can arise from dysregulated phosphorylation of the CREB family at critical junctures of somitogenesis.
Materials and Methods
Generation of ROSAAC Knock-in Mice.
A CMV/β-actin-loxP-Neo-3xpA-loxP-A-CREB-IRES-nlsEGFP cassette was knocked into the ROSA26 locus (52) to generate the ROSAAC allele (Fig. S3), which was then backcrossed to C57BL/6J for eight generations. Genotyping was performed by using standard PCR with primers listed in Table S2.
Immunostaining.
Whole-mount immunostaining protocols described previously (16) were applied to whole or half tails of CD1 (Charles River), Wnt3a+/vt, and Wnt3avt/vt (JAX) embryos at stages specified in the text. Half tails were cultured according to conditions described previously (74). Antibody dilutions were 1:50 rabbit anti–P-CREB (Cell Signaling) and 1:100 mouse anti-CREB (Cell Signaling). For immunohistochemistry on sections, tails were fixed for 25 min in 4% (wt/vol) paraformaldehyde in PBS solution and subjected to methods described previously (46) using 1:200 rabbit anti-P-CREB. Following incubation with 1:200 goat anti-rabbit IgG-HRP or 1:200 horse anti-mouse IgG-HRP (Vector), DAB reactions were used for visualization. For fluorescence, sections were processed according to a previous report (75) by using 1:100 rabbit anti–P-CREB, 1:100 rabbit anti-NICD (Cell Signaling), and 1:500 anti-fibronectin (Abcam), followed by1:1,000 Alexa 488 goat anti-rabbit IgG (Molecular Probes) or 1:200 horse anti-rabbit-HRP (Vector) and 1/50 TSA plus Cy3 (Perkin-Elmer) reaction. For actin, 1 U/μL of Alexa 568 phalloidin (Molecular Probes) was used, and, for nuclei, DAPI (1 μg/mL) was used.
WISH.
Antisense RNA probes used were as follows: Axin2, Dll-1, Mesp2, Paraxis, Uncx4.1, and Wnt3a. Hes7 (gift from R. Kageyama, Kyoto University, Kyoto, Japan), Lfng (gift from T. Gridley, Maine Medical Center Research Institute, Scarborough, ME), Ripply2, Tbx6, and Tbx18 (gifts from T. Yamaguchi, National Cancer Institute, Frederick, MD) were used for standard WISH (46). Most genes were repeated four times, whereas cycling genes were repeated at least five times for each genotype.
Histology.
H&E stain was applied to sections (60), whereas Alizarin red and Alcian blue were used for whole-mount skeletal staining (76).
Microarray.
E8.75 region II PSMs from ROSAAC/AC or ROSAAC/AC;T-Cre embryos were used for cRNA synthesis: three embryos per replicate and three replicates for each genotype. Affymetrix Mouse 430 2.0 GeneChips were used (subcontracted to Yale Center for Genome Analysis). Expression console software version 1.1 (Affymetrix) was used for chip scanning, data quality assessment, and normalization. DNA ARRAY 4.0 software and the Student t test were applied to identify differentially expressed genes by the criteria of ≥1.5 fold change and P ≤ 0.05. Microarray data was deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE46426).
μChIP.
Region II PSMs (n = 20–25) were dissected from E9.5 CD1s and prepared for µChIP analyses as described previously (77, 78) by using 2.4 µg of P-CREB antibodies (Millipore) or rabbit IgG (Millipore). µChIP samples were applied to SYBR green real-time PCR (Qiagen) by using an Opticon instrument (MJ Research). PCR primers for various CRE-sites are listed in Table S2. Occupancy of P-CREB to CRE-sites was shown as percent input (amount of ChIP DNA/amount of input DNA × 100) after subtracting IgG controls.
Supplementary Material
Acknowledgments
We thank the members of the laboratory of C.-M.F. for reading the manuscript; Samantha Satchell, Evan Siple, and Eugenia Dikovskaia for technical assistance; Dr. Alice Chen for initial observation of the segmentation defect in rare CREB mutants; and Drs. David Ginty and Terry Yamaguchi for comments and reagents. This work was supported by the Carnegie Endowment and National Institutes of Health Grants HD035596 and HD035596-S (to C.-M.F. and T.P.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data reported in this paper has been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE46426).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222115110/-/DCSupplemental.
References
- 1.Tam PPL. The control of somitogenesis in mouse embryos. J Embryol Exp Morphol. 1981;65(Suppl):103–128. [PubMed] [Google Scholar]
- 2.Kulesa PM, Fraser SE. Cell dynamics during somite boundary formation revealed by time-lapse analysis. Science. 2002;298(5595):991–995. doi: 10.1126/science.1075544. [DOI] [PubMed] [Google Scholar]
- 3.Christ B, Ordahl CP. Early stages of chick somite development. Anat Embryol (Berl) 1995;191(5):381–396. doi: 10.1007/BF00304424. [DOI] [PubMed] [Google Scholar]
- 4.Pourquié O, Tam PP. A nomenclature for prospective somites and phases of cyclic gene expression in the presomitic mesoderm. Dev Cell. 2001;1(5):619–620. doi: 10.1016/s1534-5807(01)00082-x. [DOI] [PubMed] [Google Scholar]
- 5.Aoyama H, Asamoto K. The developmental fate of the rostral/caudal half of a somite for vertebra and rib formation: Experimental confirmation of the resegmentation theory using chick-quail chimeras. Mech Dev. 2000;99(1-2):71–82. doi: 10.1016/s0925-4773(00)00481-0. [DOI] [PubMed] [Google Scholar]
- 6.Smith LJ, Nerurkar NL, Choi KS, Harfe BD, Elliott DM. Degeneration and regeneration of the intervertebral disc: lessons from development. Dis Models Mech. 2011;4(1):31–41. doi: 10.1242/dmm.006403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bronner-Fraser M. Analysis of the early stages of trunk neural crest migration in avian embryos using monoclonal antibody HNK-1. Dev Biol. 1986;115(1):44–55. doi: 10.1016/0012-1606(86)90226-5. [DOI] [PubMed] [Google Scholar]
- 8.Keynes RJ, Stern CD. Segmentation in the vertebrate nervous system. Nature. 1984;310(5980):786–789. doi: 10.1038/310786a0. [DOI] [PubMed] [Google Scholar]
- 9.Dequéant ML, Pourquié O. Segmental patterning of the vertebrate embryonic axis. Nat Rev Genet. 2008;9(5):370–382. doi: 10.1038/nrg2320. [DOI] [PubMed] [Google Scholar]
- 10.Cooke J, Zeeman EC. A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J Theor Biol. 1976;58(2):455–476. doi: 10.1016/s0022-5193(76)80131-2. [DOI] [PubMed] [Google Scholar]
- 11.Dubrulle J, McGrew MJ, Pourquié O. FGF signaling controls somite boundary position and regulates segmentation clock control of spatiotemporal Hox gene activation. Cell. 2001;106(2):219–232. doi: 10.1016/s0092-8674(01)00437-8. [DOI] [PubMed] [Google Scholar]
- 12.Aulehla A, et al. Wnt3a plays a major role in the segmentation clock controlling somitogenesis. Dev Cell. 2003;4(3):395–406. doi: 10.1016/s1534-5807(03)00055-8. [DOI] [PubMed] [Google Scholar]
- 13.Naiche LA, Holder N, Lewandoski M. FGF4 and FGF8 comprise the wavefront activity that controls somitogenesis. Proc Natl Acad Sci USA. 2011;108(10):4018–4023. doi: 10.1073/pnas.1007417108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunty WC, Jr, et al. Wnt3a/beta-catenin signaling controls posterior body development by coordinating mesoderm formation and segmentation. Development. 2008;135(1):85–94. doi: 10.1242/dev.009266. [DOI] [PubMed] [Google Scholar]
- 15.Wahl MB, Deng C, Lewandoski M, Pourquié O. FGF signaling acts upstream of the NOTCH and WNT signaling pathways to control segmentation clock oscillations in mouse somitogenesis. Development. 2007;134(22):4033–4041. doi: 10.1242/dev.009167. [DOI] [PubMed] [Google Scholar]
- 16.Sawada A, et al. Fgf/MAPK signalling is a crucial positional cue in somite boundary formation. Development. 2001;128(23):4873–4880. doi: 10.1242/dev.128.23.4873. [DOI] [PubMed] [Google Scholar]
- 17.Saga Y, Takeda H. The making of the somite: Molecular events in vertebrate segmentation. Nat Rev Genet. 2001;2(11):835–845. doi: 10.1038/35098552. [DOI] [PubMed] [Google Scholar]
- 18.Aulehla A, et al. A beta-catenin gradient links the clock and wavefront systems in mouse embryo segmentation. Nat Cell Biol. 2008;10(2):186–193. doi: 10.1038/ncb1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang YJ, et al. Notch signalling and the synchronization of the somite segmentation clock. Nature. 2000;408(6811):475–479. doi: 10.1038/35044091. [DOI] [PubMed] [Google Scholar]
- 20.Delaune EA, François P, Shih NP, Amacher SL. Single-cell-resolution imaging of the impact of Notch signaling and mitosis on segmentation clock dynamics. Dev Cell. 2012;23(5):995–1005. doi: 10.1016/j.devcel.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okubo Y, et al. Lfng regulates the synchronized oscillation of the mouse segmentation clock via trans-repression of Notch signalling. Nat Commun. 2012;3:1141. doi: 10.1038/ncomms2133. [DOI] [PubMed] [Google Scholar]
- 22.Hofmann M, et al. WNT signaling, in synergy with T/TBX6, controls Notch signaling by regulating Dll1 expression in the presomitic mesoderm of mouse embryos. Genes Dev. 2004;18(22):2712–2717. doi: 10.1101/gad.1248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morimoto M, Takahashi Y, Endo M, Saga Y. The Mesp2 transcription factor establishes segmental borders by suppressing Notch activity. Nature. 2005;435(7040):354–359. doi: 10.1038/nature03591. [DOI] [PubMed] [Google Scholar]
- 24.Bessho Y, et al. Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev. 2001;15(20):2642–2647. doi: 10.1101/gad.930601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dale JK, et al. Periodic notch inhibition by lunatic fringe underlies the chick segmentation clock. Nature. 2003;421(6920):275–278. doi: 10.1038/nature01244. [DOI] [PubMed] [Google Scholar]
- 26.Oginuma M, et al. The oscillation of Notch activation, but not its boundary, is required for somite border formation and rostral-caudal patterning within a somite. Development. 2010;137(9):1515–1522. doi: 10.1242/dev.044545. [DOI] [PubMed] [Google Scholar]
- 27.Yasuhiko Y, et al. Tbx6-mediated Notch signaling controls somite-specific Mesp2 expression. Proc Natl Acad Sci USA. 2006;103(10):3651–3656. doi: 10.1073/pnas.0508238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oginuma M, Niwa Y, Chapman DL, Saga Y. Mesp2 and Tbx6 cooperatively create periodic patterns coupled with the clock machinery during mouse somitogenesis. Development. 2008;135(15):2555–2562. doi: 10.1242/dev.019877. [DOI] [PubMed] [Google Scholar]
- 29.Morimoto M, et al. The negative regulation of Mesp2 by mouse Ripply2 is required to establish the rostro-caudal patterning within a somite. Development. 2007;134(8):1561–1569. doi: 10.1242/dev.000836. [DOI] [PubMed] [Google Scholar]
- 30.Takahashi J, et al. Analysis of Ripply1/2-deficient mouse embryos reveals a mechanism underlying the rostro-caudal patterning within a somite. Dev Biol. 2010;342(2):134–145. doi: 10.1016/j.ydbio.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 31.Foulkes NS, Borrelli E, Sassone-Corsi P. CREM gene: Use of alternative DNA-binding domains generates multiple antagonists of cAMP-induced transcription. Cell. 1991;64(4):739–749. doi: 10.1016/0092-8674(91)90503-q. [DOI] [PubMed] [Google Scholar]
- 32.Montminy MR, Bilezikjian LM. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature. 1987;328(6126):175–178. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- 33.Lee KA, et al. A cellular protein, activating transcription factor, activates transcription of multiple E1A-inducible adenovirus early promoters. Proc Natl Acad Sci USA. 1987;84(23):8355–8359. doi: 10.1073/pnas.84.23.8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hurst HC, Jones NC. Identification of factors that interact with the E1A-inducible adenovirus E3 promoter. Genes Dev. 1987;1(10):1132–1146. doi: 10.1101/gad.1.10.1132. [DOI] [PubMed] [Google Scholar]
- 35.Vinson C, et al. Classification of human B-ZIP proteins based on dimerization properties. Mol Cell Biol. 2002;22(18):6321–6335. doi: 10.1128/MCB.22.18.6321-6335.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamamoto KK, Gonzalez GA, Biggs WH, 3rd, Montminy MR. Phosphorylation-induced binding and transcriptional efficacy of nuclear factor CREB. Nature. 1988;334(6182):494–498. doi: 10.1038/334494a0. [DOI] [PubMed] [Google Scholar]
- 37.Hai TW, Liu F, Coukos WJ, Green MR. Transcription factor ATF cDNA clones: An extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 1989;3(12B):2083–2090. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- 38.Laoide BM, Foulkes NS, Schlotter F, Sassone-Corsi P. The functional versatility of CREM is determined by its modular structure. EMBO J. 1993;12(3):1179–1191. doi: 10.1002/j.1460-2075.1993.tb05759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loriaux MM, Brennan RG, Goodman RH. Modulatory function of CREB.CREM alpha heterodimers depends upon CREM alpha phosphorylation. J Biol Chem. 1994;269(46):28839–28843. [PubMed] [Google Scholar]
- 40.Hurst HC, Masson N, Jones NC, Lee KA. The cellular transcription factor CREB corresponds to activating transcription factor 47 (ATF-47) and forms complexes with a group of polypeptides related to ATF-43. Mol Cell Biol. 1990;10(12):6192–6203. doi: 10.1128/mcb.10.12.6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hurst HC, Totty NF, Jones NC. Identification and functional characterisation of the cellular activating transcription factor 43 (ATF-43) protein. Nucleic Acids Res. 1991;19(17):4601–4609. doi: 10.1093/nar/19.17.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rehfuss RP, Walton KM, Loriaux MM, Goodman RH. The cAMP-regulated enhancer-binding protein ATF-1 activates transcription in response to cAMP-dependent protein kinase A. J Biol Chem. 1991;266(28):18431–18434. [PubMed] [Google Scholar]
- 43.de Groot RP, den Hertog J, Vandenheede JR, Goris J, Sassone-Corsi P. Multiple and cooperative phosphorylation events regulate the CREM activator function. EMBO J. 1993;12(10):3903–3911. doi: 10.1002/j.1460-2075.1993.tb06068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2(8):599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 45.Shaywitz AJ, Greenberg ME. CREB: A stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. erratum (2003) 72:vii. [DOI] [PubMed] [Google Scholar]
- 46.Chen AE, Ginty DD, Fan CM. Protein kinase A signalling via CREB controls myogenesis induced by Wnt proteins. Nature. 2005;433(7023):317–322. doi: 10.1038/nature03126. [DOI] [PubMed] [Google Scholar]
- 47.Ginty DD, et al. Regulation of CREB phosphorylation in the suprachiasmatic nucleus by light and a circadian clock. Science. 1993;260(5105):238–241. doi: 10.1126/science.8097062. [DOI] [PubMed] [Google Scholar]
- 48.Hagiwara M, et al. Coupling of hormonal stimulation and transcription via the cyclic AMP-responsive factor CREB is rate limited by nuclear entry of protein kinase A. Mol Cell Biol. 1993;13(8):4852–4859. doi: 10.1128/mcb.13.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bleckmann SC, et al. Activating transcription factor 1 and CREB are important for cell survival during early mouse development. Mol Cell Biol. 2002;22(6):1919–1925. doi: 10.1128/MCB.22.6.1919-1925.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahn S, et al. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol Cell Biol. 1998;18(2):967–977. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Acharya A, Rishi V, Moll J, Vinson C. Experimental identification of homodimerizing B-ZIP families in Homo sapiens. J Struct Biol. 2006;155(2):130–139. doi: 10.1016/j.jsb.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 52.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 53.Perantoni AO, et al. Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development. 2005;132(17):3859–3871. doi: 10.1242/dev.01945. [DOI] [PubMed] [Google Scholar]
- 54.Burgess R, Rawls A, Brown D, Bradley A, Olson EN. Requirement of the paraxis gene for somite formation and musculoskeletal patterning. Nature. 1996;384(6609):570–573. doi: 10.1038/384570a0. [DOI] [PubMed] [Google Scholar]
- 55.Choi KS, Cohn MJ, Harfe BD. Identification of nucleus pulposus precursor cells and notochordal remnants in the mouse: Implications for disk degeneration and chordoma formation. Dev Dyn. 2008;237(12):3953–3958. doi: 10.1002/dvdy.21805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leitges M, Neidhardt L, Haenig B, Herrmann BG, Kispert A. The paired homeobox gene Uncx4.1 specifies pedicles, transverse processes and proximal ribs of the vertebral column. Development. 2000;127(11):2259–2267. doi: 10.1242/dev.127.11.2259. [DOI] [PubMed] [Google Scholar]
- 57.Bussen M, et al. The T-box transcription factor Tbx18 maintains the separation of anterior and posterior somite compartments. Genes Dev. 2004;18(10):1209–1221. doi: 10.1101/gad.300104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang X, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci USA. 2005;102(12):4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dequéant ML, et al. A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science. 2006;314(5805):1595–1598. doi: 10.1126/science.1133141. [DOI] [PubMed] [Google Scholar]
- 60.Hrabĕ de Angelis M, McIntyre J, 2nd, Gossler A. Maintenance of somite borders in mice requires the Delta homologue DII1. Nature. 1997;386(6626):717–721. doi: 10.1038/386717a0. [DOI] [PubMed] [Google Scholar]
- 61.Takahashi Y, Inoue T, Gossler A, Saga Y. Feedback loops comprising Dll1, Dll3 and Mesp2, and differential involvement of Psen1 are essential for rostrocaudal patterning of somites. Development. 2003;130(18):4259–4268. doi: 10.1242/dev.00629. [DOI] [PubMed] [Google Scholar]
- 62.Beckers J, et al. Distinct regulatory elements direct delta1 expression in the nervous system and paraxial mesoderm of transgenic mice. Mech Dev. 2000;95(1-2):23–34. doi: 10.1016/s0925-4773(00)00322-1. [DOI] [PubMed] [Google Scholar]
- 63.Chan T, et al. Ripply2 is essential for precise somite formation during mouse early development. FEBS Lett. 2007;581(14):2691–2696. doi: 10.1016/j.febslet.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 64.Yasuhiko Y, et al. Functional importance of evolutionally conserved Tbx6 binding sites in the presomitic mesoderm-specific enhancer of Mesp2. Development. 2008;135(21):3511–3519. doi: 10.1242/dev.027144. [DOI] [PubMed] [Google Scholar]
- 65.Hagiwara M, et al. Transcriptional attenuation following cAMP induction requires PP-1-mediated dephosphorylation of CREB. Cell. 1992;70(1):105–113. doi: 10.1016/0092-8674(92)90537-m. [DOI] [PubMed] [Google Scholar]
- 66.Diez del Corral R, et al. Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron. 2003;40(1):65–79. doi: 10.1016/s0896-6273(03)00565-8. [DOI] [PubMed] [Google Scholar]
- 67.Moreno TA, Kintner C. Regulation of segmental patterning by retinoic acid signaling during Xenopus somitogenesis. Dev Cell. 2004;6(2):205–218. doi: 10.1016/s1534-5807(04)00026-7. [DOI] [PubMed] [Google Scholar]
- 68.Hummler E, et al. Targeted mutation of the CREB gene: compensation within the CREB/ATF family of transcription factors. Proc Natl Acad Sci USA. 1994;91(12):5647–5651. doi: 10.1073/pnas.91.12.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Impey S, et al. Defining the CREB regulon: A genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119(7):1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 70.Takahashi Y, et al. Transcription factors Mesp2 and Paraxis have critical roles in axial musculoskeletal formation. Dev Dyn. 2007;236(6):1484–1494. doi: 10.1002/dvdy.21178. [DOI] [PubMed] [Google Scholar]
- 71.Whittock NV, et al. Mutated MESP2 causes spondylocostal dysostosis in humans. Am J Hum Genet. 2004;74(6):1249–1254. doi: 10.1086/421053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sparrow DB, et al. Mutation of the LUNATIC FRINGE gene in humans causes spondylocostal dysostosis with a severe vertebral phenotype. Am J Hum Genet. 2006;78(1):28–37. doi: 10.1086/498879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bulman MP, et al. Mutations in the human delta homologue, DLL3, cause axial skeletal defects in spondylocostal dysostosis. Nat Genet. 2000;24(4):438–441. doi: 10.1038/74307. [DOI] [PubMed] [Google Scholar]
- 74.Correia KM, Conlon RA. Surface ectoderm is necessary for the morphogenesis of somites. Mech Dev. 2000;91(1-2):19–30. doi: 10.1016/s0925-4773(99)00260-9. [DOI] [PubMed] [Google Scholar]
- 75.Lopez TP, Fan CM. A transgenic Tbx6;CreER(T2) line for inducible gene manipulation in the presomitic mesoderm. Genesis. 2012;50(6):490–495. doi: 10.1002/dvg.20827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Martinelli DC, Fan CM. Gas1 extends the range of Hedgehog action by facilitating its signaling. Genes Dev. 2007;21(10):1231–1243. doi: 10.1101/gad.1546307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vilhais-Neto GC, et al. Rere controls retinoic acid signalling and somite bilateral symmetry. Nature. 2010;463(7283):953–957. doi: 10.1038/nature08763. [DOI] [PubMed] [Google Scholar]
- 78.Dahl JA, Collas P. A rapid micro chromatin immunoprecipitation assay (microChIP) Nat Protoc. 2008;3(6):1032–1045. doi: 10.1038/nprot.2008.68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.