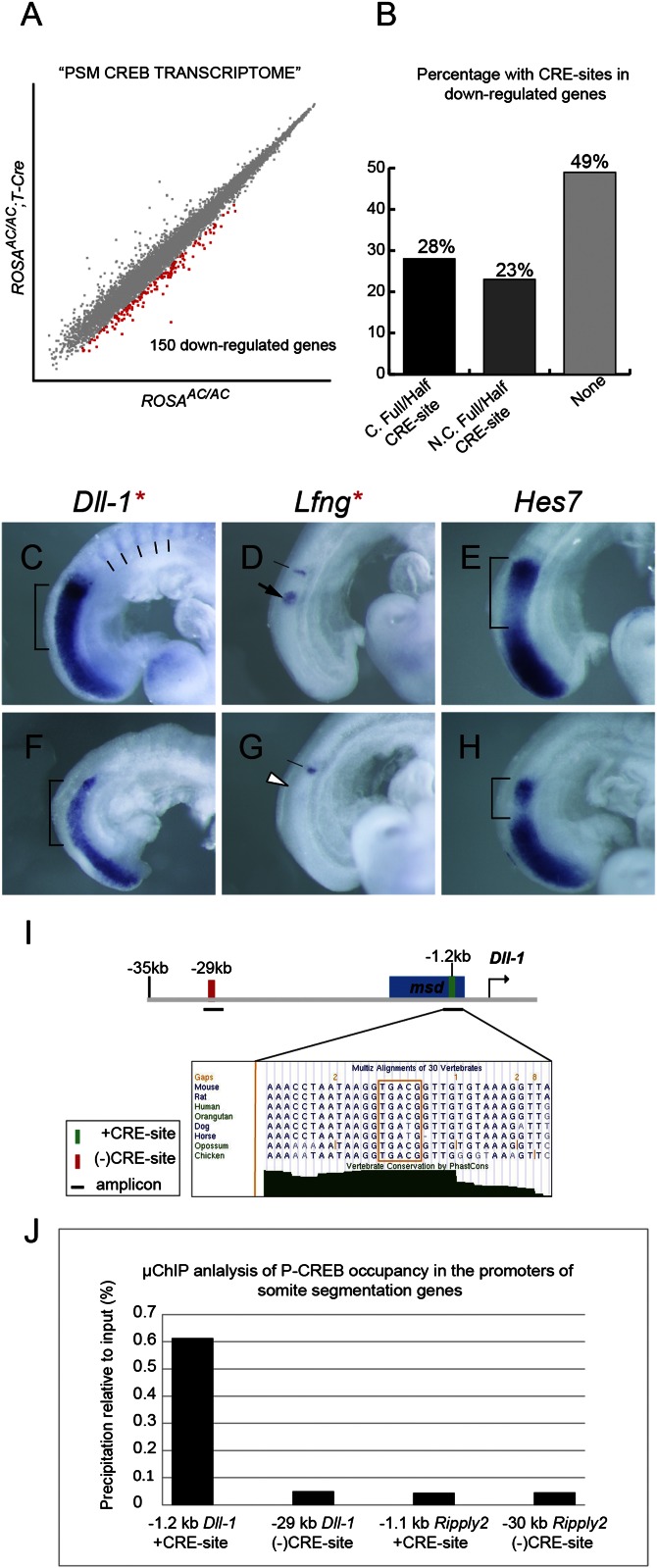
Fig. 5.
The CREB family controls somite morphogenesis through Dll-1/Notch signaling. (A) Scatter plot of log2-transformed hybridization signal intensity values between ROSAAC/AC;T-Cre (y axis) mutants, and ROSAAC/AC (x axis) controls. A total of 150 genes (red squares) were found to be down-regulated. (B) Bar graph tabulates percentage of down-regulated genes with conserved (marked as “C”) full/half CRE-sites, nonconserved (N.C.) full/half CRE-sites, and none. (C–H) WISH analysis of Dll-1 (C and F), Lfng (D and G), and Hes7 (E and H) in controls (C–E) and mutants (F–H): brackets indicate altered Dll-1 levels and Hes7 domains between controls and mutants; lines in control (C) marks Dll-1 expression in posterior somite halves; white arrowhead in mutant (G) indicates strongly reduced Lfng cyclic expression; red asterisks indicate genes with a putative CRE-site. (I) Diagram of the Dll-1 promoter and −35 kb upstream of the transcriptional start site. Blue box is the msd enhancer. Magnified box is a multiple sequence alignment of a conserved +CRE-site (orange box); green bar indicates CRE-site; red bar indicates (−)CRE-site; solid lines indicate amplicons. (J) Bar graph of μChIP quantification. P-CREB occupation at the −1.2 kb CRE-site (0.614%) in the Dll-1 promoter is 12.3-fold higher than the (−)CRE-site control (0.050%; −29 kb). P-CREB binding to the −1.1 kb CRE-site (0.044%) in the Ripply2 promoter is comparable to the (−)CRE-site control (0.046%; −30 kb).