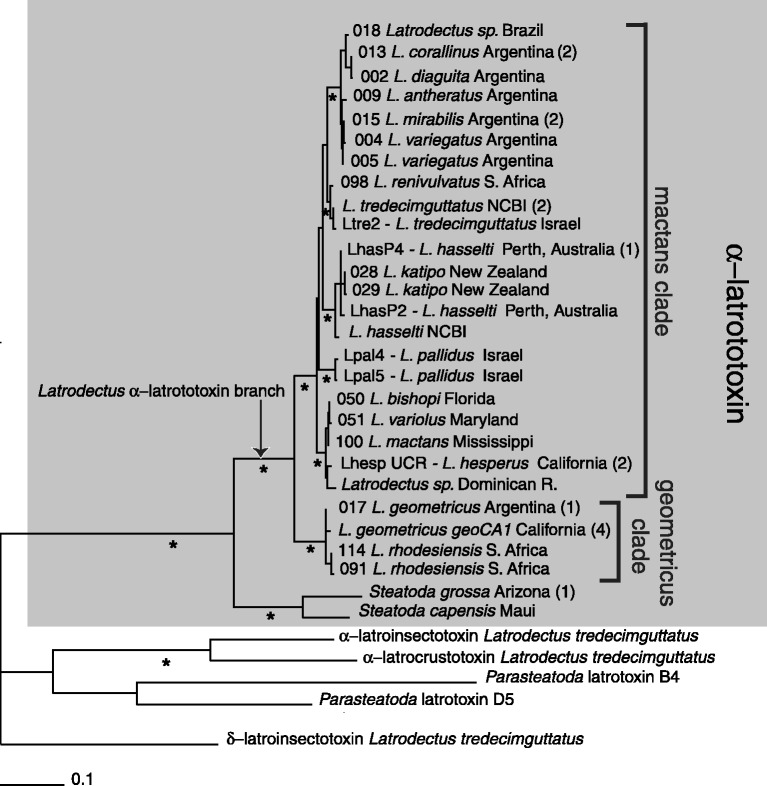
Fig. 4.
ML phylogram from conserved 642 bp alignment (618 bp PCR fragment) of α-latrotoxin and latrotoxin homologs. Asterisks indicate nodes supported by ≥88% bootstrap support from 100 replicates (ML) or 1,000 replicates (parsimony), and ≥0.95 clade posterior support values in partitioned Bayesian analysis. Numbers in parentheses indicate numbers of additional identical sequences (supplementary table S1, Supplementary Material online: 013 Latrodectus corallinus identical to 020 L. corallinus and 003 L. diaguita; 015 L. mirabilis identical to 014 L. mirabilis and 010 Latrodectus sp.; L. hasselti LhasP4 identical to LhasP5; L. tredecimguttatus NCBI identical to 036 L. tredecimguttatus and Ltre4; L. hesperus LhespUCR identical to Lhesmar1 and 049 L. hesperus; 017 L. geometricus identical to 016 L. geometricus; L. geometricus geoCA1 identical to 034 L. geometricus, 033 L. geometricus, 097 L. geometricus and 099 L. geometricus; S. grossa Arizona identical to S. grossa Maui). The tree was rooted with the latrotoxin paralog δ-latroinsectotoxin. Partitioned Bayesian and parsimony consensus trees for these data are shown in supplementary figures 6 and 7, Supplementary Material online, respectively.