Fig. 3.
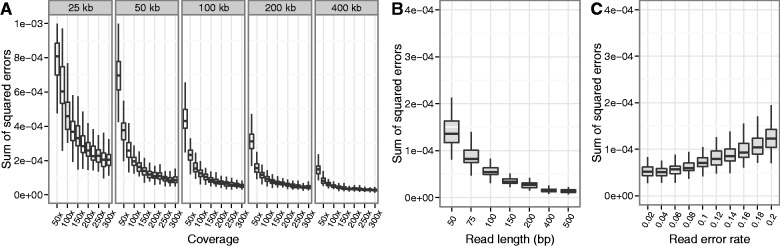
Performance of the EM algorithm increases with coverage, region width, and read length and is robust to sequencing errors. (A) Performance of the EM algorithm increases with both coverage and width of the region used for the estimation. (B) The EM algorithm performs better with longer reads, which provide more haplotype information. (C) The EM algorithm maintains good performance with increasing sequence read error rate. Empirical error rates were found to be in the range of 0.05–0.07 errors per base call. In all simulations, we simulated paired-end pooled sequence data from 162 haplotypes at randomly drawn frequencies, with 100 replicates per parameter value level. Nonvarying parameters were held at fixed values representative of our experimental data (read length 100 bp, read error rate 0.06, coverage 200×, and region width 200 kb).
