Fig. 7.
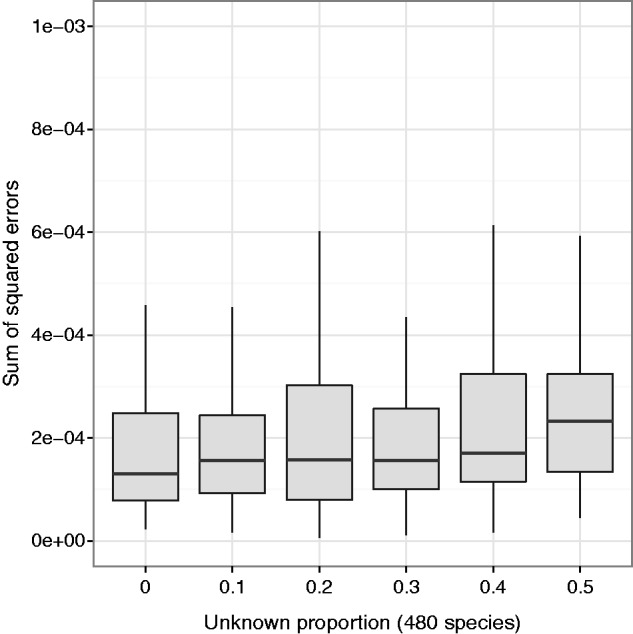
Large numbers of unknown unrelated species do not significantly affect the estimate of within-genus species frequencies. The EM algorithm with haplotype likelihood filter was run on simulated 75-bp single-end 16S sequence reads from 500 species (20 Clostridium species [known] and 480 non-Clostridium species [unknown]). “Unknown proportion” is the total proportion of reads coming uniformly at random from the 480 unknown species, with the remainder of the reads coming from the 20 known species (100× pooled coverage, 100 replicates for each unknown proportion level).
