Fig. 8.
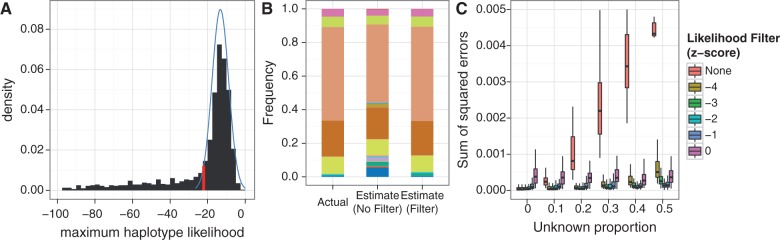
Sequence reads from unknown unrelated species push frequency estimates toward the uniform distribution; filtering the reads based on haplotype likelihood minimizes this effect. (A) Sequence reads from unknown unrelated species have low maximum haplotype likelihoods, giving rise to the long left tail of the distribution. By calculating the theoretical distribution (blue) of maximum haplotype likelihoods based on the base quality scores from the sequence data, reads whose maximum haplotype likelihood falls below a specified threshold (red, z-score threshold = −2 in this example) can be filtered out. (B) A typical example of this effect, with 50% of the reads coming from the unknown species. (C) Without filtering on the haplotype likelihoods, error in frequency estimates increases with higher proportion of unknown sequence reads. Seventy-five-base-pair single-end 16S sequence reads were simulated from 20 species, with varying proportions of reads from an unknown unrelated species (100× pooled coverage, 100 replicates per unknown proportion level).
