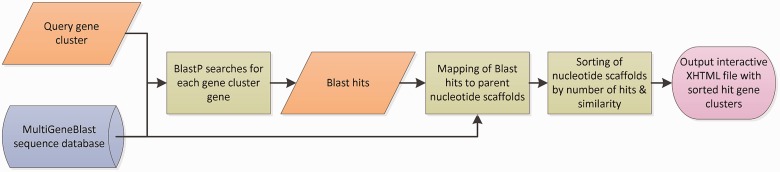
Fig. 1.
Outline of the homology search process by MultiGeneBlast. First, the amino acid translation of each gene sequence within the query gene cluster is searched against the selected MultiGeneBlast database, yielding a data set of BLAST hits. The BLAST hits are then mapped to their parent nucleotide scaffolds, based on the information from the database. The nucleotide scaffolds are then sorted according to their empirical similarity scores with the query gene cluster. Finally, the sorted list of genomic loci is displayed in an interactive XHTML file that can be viewed with any modern web browser.