Abstract
Recombination suppression leads to the structural and functional differentiation of sex chromosomes and is thus a crucial step in the process of sex chromosome evolution. Despite extensive theoretical work, the exact processes and mechanisms of recombination suppression and differentiation are not well understood. In threespine sticklebacks (Gasterosteus aculeatus), a different sex chromosome system has recently evolved by a fusion between the Y chromosome and an autosome in the Japan Sea lineage, which diverged from the ancestor of other lineages approximately 2 Ma. We investigated the evolutionary dynamics and differentiation processes of sex chromosomes based on comparative analyses of these divergent lineages using 63 microsatellite loci. Both chromosome-wide differentiation patterns and phylogenetic inferences with X and Y alleles indicated that the ancestral sex chromosomes were extensively differentiated before the divergence of these lineages. In contrast, genetic differentiation appeared to have proceeded only in a small region of the neo-sex chromosomes. The recombination maps constructed for the Japan Sea lineage indicated that recombination has been suppressed or reduced over a large region spanning the ancestral and neo-sex chromosomes. Chromosomal regions exhibiting genetic differentiation and suppressed or reduced recombination were detected continuously and sequentially in the neo-sex chromosomes, suggesting that differentiation has gradually spread from the fusion point following the extension of recombination suppression. Our study illustrates an ongoing process of sex chromosome differentiation, providing empirical support for the theoretical model postulating that recombination suppression and differentiation proceed in a gradual manner in the very early stage of sex chromosome evolution.
Keywords: chromosome fusion, Gasterosteus, neo-sex chromosome, recombination suppression, sex chromosome, sex chromosome evolution
Introduction
Theoretical studies have extensively hypothesized that sex chromosomes evolved from an ordinary pair of autosomes that acquired a sex-determining role (Ohno 1969; Rice 1996; Charlesworth et al. 2005; Bachtrog 2006a). According to a widely accepted model, recombination between sex chromosomes is initially restricted in a limited region surrounding the sex-determining gene, and recombination suppression extends along the entire chromosomes—apart from short pseudoautosomal regions. The lack of recombination allows the accumulation of deleterious mutations in the sex-limited chromosome, which in turn leads to the loss of function of genes in the nonrecombining region of this chromosome. Therefore, the suppression of recombination is a crucial step in the process of sex chromosome evolution. Two main hypotheses based on gradual and stepwise models have been proposed to explain the processes of recombination suppression (Rice 1996; Charlesworth et al. 2005), which is thought to occur through several mechanisms and ultimate causes such as sexually antagonistic selection, chromosome inversion, or other chromosomal rearrangements (Fisher 1931; Charlesworth and Hartl 1978; Bull 1983; Rice 1987; Lahn and Page 1999; Charlesworth et al. 2005). Despite extensive theoretical studies on sex chromosome evolution, the exact mechanisms and processes of recombination suppression and differentiation are not well understood.
The role of inversions in suppressing recombination between sex chromosomes has been highlighted in empirical studies of several model organisms (Lahn and Page 1999; Lemaitre et al. 2009; Wilson and Makova 2009). These studies implied that distinct clusters of sex chromosomes have suppressed recombination independently at different times, potentially via a series of Y chromosome inversions, which prevent crossing over due to the lack of chromosomal homology (Navarro et al. 1997; Andolfatto et al. 2001). For instance, the human X chromosome exhibits at least four evolutionary strata, representing distinct evolutionary histories of recombination suppression (Lahn and Page1999; Skaletsky et al. 2003; Ross et al. 2005; but see e.g., Katsura et al. 2012). Similarly, evolutionary strata have been identified in the sex chromosomes of the mouse (Sandstedt and Tucker 2004), cat (Pearks Wilkerson et al. 2008), cattle (Van Laere et al. 2008), and chicken (Handley et al. 2004; Nam and Ellegren 2008). Because therian and avian sex chromosomes arose up to approximately 200 Ma, their Y or W chromosomes retain only a few active genes and consist mainly of repetitive DNA elements, exhibiting only few clues about their autosomal origin or the processes that resulted in their degeneration (Charlesworth B and Charlesworth D 2000). Therefore, it is not certain whether inversions are the cause of recombination suppression or whether inversions occur as a consequence of ceased recombination (Charlesworth et al. 2005; Wimmer et al. 2005; Wilson and Makova 2009). In the case of the plant Silene latifolia, inversions are not actually associated with the formation of evolutionary strata; the progression of sex chromosome differentiation is gradual rather than a result of large chromosomal rearrangements (Bergero et al. 2007, 2008). Given a potential difference in the evolutionary dynamics of sex chromosomes between taxa—particularly between animals and plants (Bachtrog 2011)—more studies on several independently evolving Y chromosomes from a wider taxonomic perspective are greatly needed (Wilson and Makova 2009).
Molecular differentiation of sex chromosomes has been extensively studied using model organisms, mainly focusing on degeneration patterns in paralogous gene sequences shared between the sex chromosomes (Wilson and Makova 2009; Ellegren 2011). Recent molecular genetic and cytological analyses have shown diverse evolutionary histories and stages of sex chromosomes across different taxa (Charlesworth and Mank 2010; Janousek and Mrackova 2010; Otto et al. 2011). Unlike mammals or birds, several lower vertebrates are known to exhibit a rapid turnover of sex chromosomes, which could be achieved by different mechanisms such as the transposition of an existing sex-determining gene to an autosome, the development of a new sex-determining gene on an autosome, or by fusions between an ancestral sex chromosome and an autosome (Woram et al. 2003; Takehana et al. 2007; Cnaani et al. 2008; Ross et al. 2009). Because evolutionarily younger sex chromosomes are expected to be less degenerated, organisms with recently evolved sex chromosomes are suitable to address the initial phases of sex chromosome evolution and thus can be better used to test predicted models for the processes and mechanisms of recombination suppression and differentiation (Bachtrog 2006a). In addition, molecular processes of recombination suppression and differentiation can be assessed with recently formed neo-sex chromosomes, in which the newly fused chromosomal regions are expected to be affected by the same evolutionary forces as ancestral regions (Charlesworth 1996; Steinemann M and Steinemann S 1998; Charlesworth B and Charlesworth D 2000). Studies on Drosophila showed that the neo-Y chromosomes begin to degenerate rapidly once recombination is suppressed (Bachtrog 2006b; Bachtrog et al. 2008; Zhou and Bachtrog 2012). However, little is known about the processes and paces of recombination suppression and differentiation in neo-sex chromosomes in other taxonomic groups.
Comparative analyses of related species or intraspecific variants can provide a time scale to study the process of sex chromosome evolution. Thus far, such analyses have been conducted mainly using interspecific comparisons (Nicolas et al. 2005; Kondo et al. 2009; Ross et al. 2009; Kaiser and Bachtrog 2010; Stöck et al. 2011; Katsura et al. 2012; Pala, Hasselquist, et al. 2012). Fish exhibit diverse sex determination and sex chromosome systems (Devlin and Nagahama 2002; Mank et al. 2006). Additionally, sex chromosome rearrangements have been found in several fish species (Mank and Avise 2009; Cioffi et al. 2012; Kitano and Peichel 2012). Because of the rapid turnover of fish sex chromosomes, they are presumed to be in the early stages of evolution (Charlesworth 2004; Volff et al. 2007; Kondo et al. 2009; Ross et al. 2009). In the stickleback family (Gasterosteidae), closely related species have different sex determination and sex chromosome systems (Ross et al. 2009). Moreover, in threespine sticklebacks (Gasterosteus aculeatus), a different sex chromosome system has formed as a result of a fusion between the ancestral Y chromosome and an autosome in the Japan Sea lineage (Kitano et al. 2009). This rearrangement has occurred during or after the divergence from the ancestor of other (Pacific and Atlantic) lineages, which is estimated to have occurred approximately 2 Ma (Haglund et al. 1992; Higuchi and Goto 1996). However, no chromosomal rearrangements have been found in the Pacific and Atlantic lineages, having diverged from each other within the last 1 My (O'Reilly et al. 1993; Ortí et al. 1994; Deagle et al. 1996). Because the neo-sex chromosomes of the Japan Sea lineage have recently evolved, they are expected to be at an earlier stage of differentiation than the ancestral sex chromosomes. Comparative analyses of these divergent threespine stickleback lineages provide a framework for understanding the evolutionary dynamics of both ancestral and neo-sex chromosomes, as well as the molecular process of sex chromosome differentiation on a relatively short evolutionary time frame.
A previous study found a high degree of genetic differentiation between sex chromosomes in an Atlantic population of threespine sticklebacks (Shikano et al. 2011). Given the rapid turnover of sex chromosomes in stickleback species, sex chromosomes might be evolving rapidly in threespine sticklebacks as seen in the recent formation of a neo-sex chromosome system in the Japan Sea lineage (Kitano et al. 2009). In this study, we aimed to investigate the evolutionary dynamics and differentiation processes of both ancestral and neo-sex chromosomes of threespine sticklebacks based on intraspecific comparative analyses with the divergent lineages. The ancestral and neo-sex chromosomes of this species have been identified to correspond to linkage groups (LGs) 19 and 9, respectively (Peichel et al. 2004; Kitano et al. 2009). We uncovered the physical locations of where genetic differentiation has been achieved in these sex chromosomes using a number of microsatellite markers, which are known to be sensitive in detecting the genetic differentiation and phylogeny of sex chromosomes (Schlötterer 2000; Jobling and Tyler-Smith 2003; Underhill and Kivisild 2007; Shikano et al. 2011). In addition, we explored the evolutionary history of the sex chromosomes based on a phylogenetic approach using nine globally distributed populations. Furthermore, detailed recombination maps were constructed for the ancestral and neo-sex chromosomes and compared with the physical map data to assess the process and pace of recombination suppression and differentiation. On the basis of these analyses, we tested theoretical models to explain the process of sex chromosome differentiation in the early stages of evolution. In particular, we were interested in addressing the question whether recombination suppression and differentiation proceed according to a gradual or stepwise model.
Results
Genetic Differentiation of Sex Chromosomes
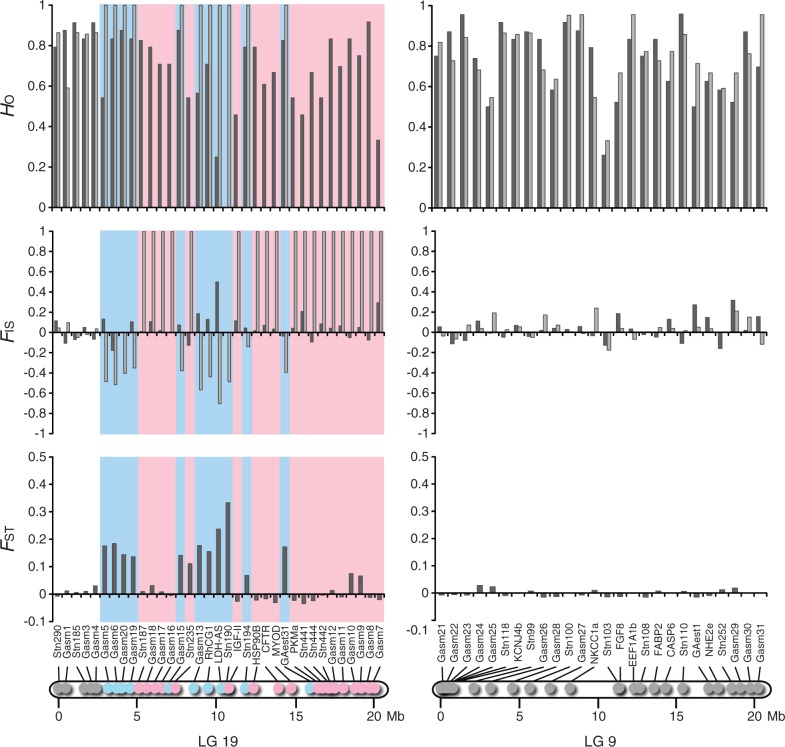
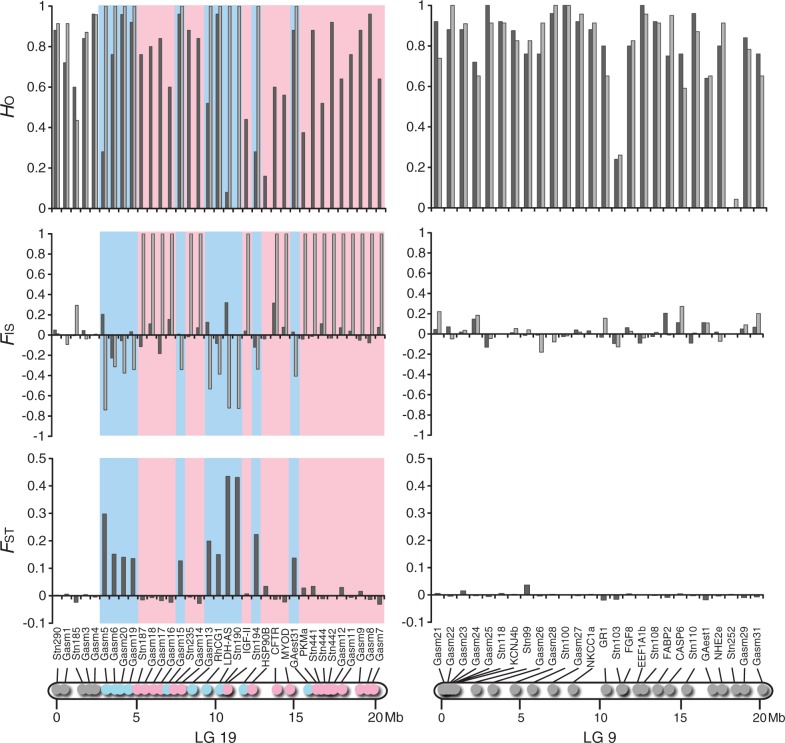
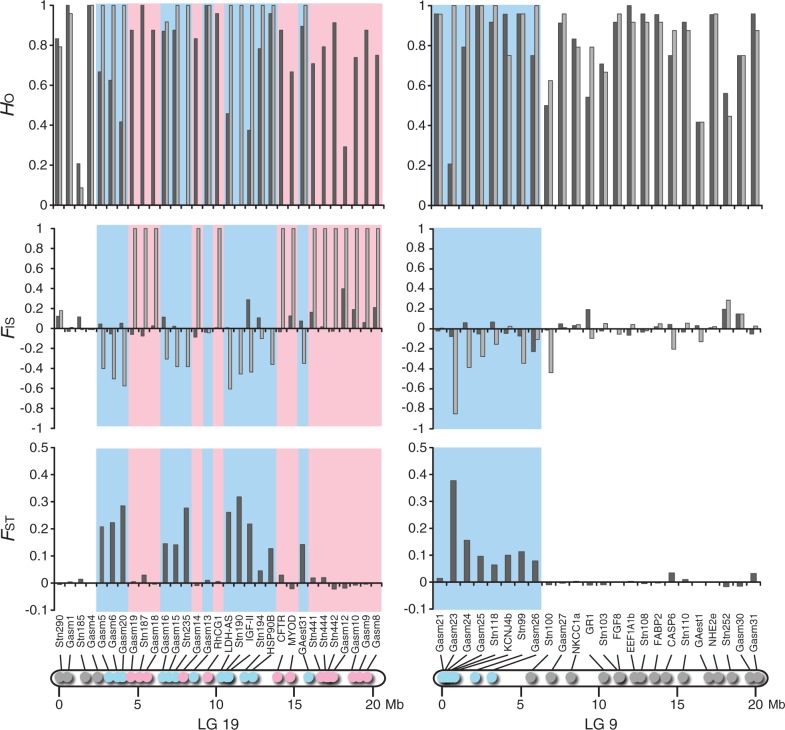
Chromosome-wide differentiation patterns in LGs 19 and 9 were assessed in three populations (P-SHI, A-HEL, and J-BIW) from the three divergent lineages using 63 loci (fig. 1 and supplementary tables S1 and S2, Supplementary Material online). In LG 19, male-specific alleles were detected at 11 of 35 loci in P-SHI and A-HEL, and 13 of 31 loci in J-BIW (figs. 2–4 and supplementary tables S3 and S4, Supplementary Material online). Most of these loci exhibited significant FST values between females and males, as well as negative FIS values in males (supplementary tables S3 and S4, Supplementary Material online). In contrast, no heterozygous individuals were found in males at 19 loci in P-SHI and A-HEL and 14 loci in J-BIW, implying male-specific null alleles (figs. 2–4 and supplementary table S3, Supplementary Material online). FIS values were significantly positive in males at all these loci except for one locus (HSP90B) in A-HEL, at which only one allele was found in males (supplementary table S3, Supplementary Material online). No sex-specific allelic patterns, as well as no differentiation between females and males, were detected at the remaining five loci in P-SHI and A-HEL, and four loci in J-BIW (figs. 2–4 and supplementary tables S3 and S4, Supplementary Material online). These loci were located in the first 2.5 Mb region of LG 19 in all populations. In the 29 loci commonly used for the analyses in the three populations, male-specific alleles and null alleles were observed at identical loci in P-SHI and A-HEL, whereas different allelic patterns were detected at six loci in J-BIW (figs. 2–4). These different patterns were found in a chromosomal region between 4.5 Mb and 12.5 Mb. All the loci located in the last 3.9 Mb region of LG 19 exhibited male-specific null alleles in the three populations (figs. 2–4). The sex-determining locus has been mapped to near this region (Peichel et al. 2004).
Fig. 1.
Sampling locations of nine study populations.
Fig. 2.
Observed heterozygosity (HO) and FIS in females (black) and males (gray) and FST between females and males at 35 loci on LG 19 (left) and at 26 loci on LG 9 (right) in P-SHI. Locus positions are indicated in the physical maps. The blue color represents the loci where male-specific alleles were observed, and the red color indicates the loci where no heterozygous males were found.
Fig. 3.
Observed heterozygosity (HO) and FIS in females (black) and males (gray) and FST between females and males at 35 loci on LG 19 (left) and at 26 loci on LG 9 (right) in A-HEL. Locus positions are indicated in the physical maps. The blue color represents the loci where male-specific alleles were observed, and the red color indicates the loci where no heterozygous males were found.
Fig. 4.
Observed heterozygosity (HO) and FIS in females (black) and males (gray) and FST between females and males at 31 loci on LG 19 (left) and at 24 loci on LG 9 (right) in J-BIW. Locus positions are indicated in the physical maps. The blue color represents the loci where male-specific alleles were observed, and the red color indicates the loci where no heterozygous males were found.
In LG 9, none of the 26 loci investigated showed significant FST between females and males or sex-specific FIS in P-SHI and A-HEL (figs. 2 and 3 and supplementary tables S3 and S4, Supplementary Material online). In J-BIW, male-specific alleles were detected at 8 of 24 loci. All these eight loci were found in the first 3.2 Mb region of LG 9, corresponding to 15.8% of the length (fig. 4 and supplementary table S4, Supplementary Material online). Of these, seven showed significant FST values between females and males and four showed negative FIS values in males (supplementary tables S3 and S4, Supplementary Material online). There was no indication of male-specific null alleles in any of the loci located on LG 9.
In LGs 19 and 9, significant linkage disequilibrium between loci was detected for 75 combinations of 1,830 in P-SHI, 43 of 1,830 in A-HEL, and 56 of 1,485 in J-BIW (supplementary tables S5–S7, Supplementary Material online). In P-SHI and A-HEL, most of the significant combinations were observed within LG 19 (supplementary tables S5 and S6, Supplementary Material online). In contrast, significant linkage disequilibrium was found in combinations between LGs 19 and 9 (18 comparisons), in addition to those within LG 19 (37 comparisons) in J-BIW (supplementary table S7, Supplementary Material online). Most of the significant combinations observed between LGs 19 and 9 occurred in comparisons with the loci having male-specific alleles in LG 9.
Phylogenetic Analyses of Sex Chromosome Evolution
The differentiation patterns of sex chromosomes were investigated in an additional six populations using 23 loci (15 in LG 19 and eight in LG 9; fig. 1 and supplementary tables S1 and S8, Supplementary Material online). In the populations of the Pacific and Atlantic lineages, male-specific alleles and significant FST between sexes were detected for the same 11 loci in LG 19 as in P-SHI and A-HEL (see previous section), except for Gasm13 in A-MAI (supplementary table S9, Supplementary Material online). Similarly, significantly high FIS values were observed in males for the same four loci in LG 19 as in P-SHI and A-HEL, implying the presence of male-specific null alleles (supplementary table S8, Supplementary Material online). No sex-specific patterns were detected for eight loci in LG 9 in any of these populations (supplementary table S8, Supplementary Material online). In the J-MAS population from the Japan Sea lineage, male-specific alleles and null alleles were indicated for the same loci as in J-BIW (see previous section; supplementary tables S8 and S9, Supplementary Material online).
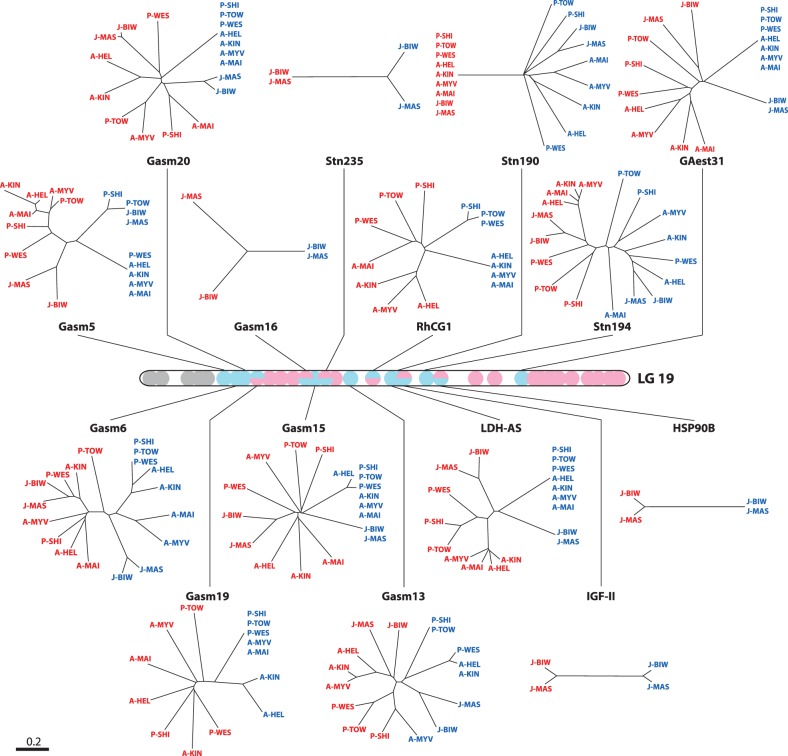
The neighbor-joining (NJ) tree of DCE distances was constructed using the 15 loci from LG 19 in which male-specific alleles were identified. Each sex chromosome was clustered together across all populations with a high bootstrap support (99%), whereas no clustering was found for X and Y chromosomes in the respective populations (fig. 5A). The NJ tree for each locus showed consistent differentiation patterns in most of the loci, independent of chromosomal location (fig. 6). Similarly, phylogenetic analyses based on eight loci in LG 9 showed that each neo-sex chromosome grouped together with a high bootstrap support (100%) in the Japan Sea lineage (fig. 5B).
Fig. 5.
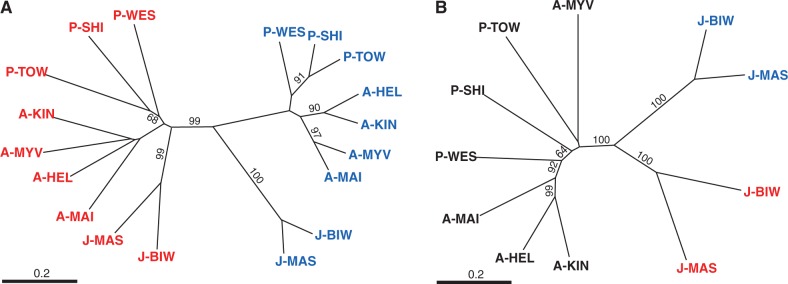
Phylogenetic trees based on DCE distances for X (red) and Y chromosomal alleles (blue) at 15 loci on LG 19 (A) and at eight loci on LG 9 (B) in nine populations. Data from males are indicated in black. Bootstrap supports above 60% are indicated.
Fig. 6.
Phylogenetic trees based on DCE distances for X (red) and Y chromosomal alleles (blue) at each locus on LG 19 in nine populations. The colors in the physical map indicate allelic patterns at 36 loci on LG 19 in P-SHI, A-HEL, and J-BIW (see figs. 2–4 for details). For the loci indicated by two colors, the upper color is based on P-SHI and A-HEL and the lower color J-BIW.
Recombination and Differentiation in Ancestral and Neo-sex Chromosomes
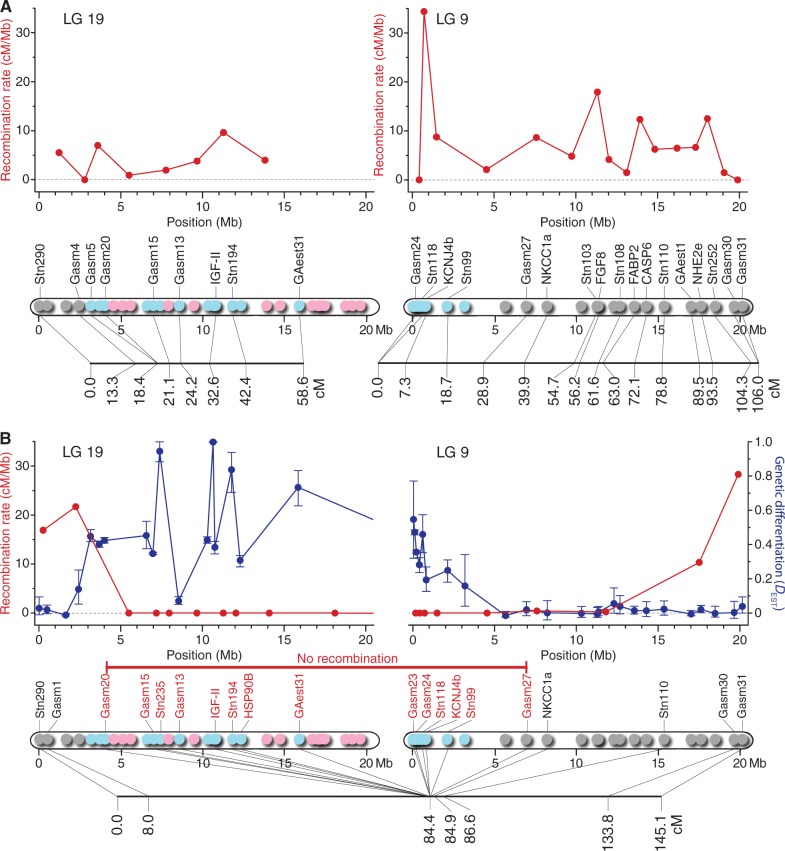
In the consensus analyses of three families in the Japan Sea lineage, two LGs corresponding to LGs 19 and 9 were identified in the female meiosis with high LOD (logarithm of the odds) scores (≥10.2) in each LG. The female-specific LG 19 map included nine loci, spanning 58.6 centimorgan (cM) with an average intermarker distance of 7.3 cM (fig. 7A). The female-specific LG 9 map consisted of 17 loci, spanning 106.0 cM with an average intermarker distance of 6.6 cM (fig. 7A). Recombination rates between adjacent loci were estimated to be 0.00–9.63 cM/Mb with an average of 4.10 cM/Mb in LG 19 and 0.00–34.38 cM/Mb with an average of 8.00 cM/Mb in LG 9 (fig. 7A).
Fig. 7.
Recombination rates (red) and genetic linkage maps for LGs 19 and 9 in female meiosis (A) and male meiosis (B) in J-BIW. DEST values and 95% confidence intervals (blue) are shown in (B). Recombination rates are plotted against intermediate physical location between adjacent loci. The numbers in the linkage maps represent genetic map distances in Kosambi centimorgan (cM). The colors in the physical maps indicate allelic patterns at 55 loci on LGs 19 and 9 in J-BIW (see fig. 4 for details).
One LG was obtained for the male meiosis with a high LOD score (56.8) between the adjacent linked loci of LGs 19 and 9. The male-specific linkage map comprised 20 loci, spanning 145.0 cM with intermarker distances of 0.0–76.3 cM (fig. 7B). Recombination rates between adjacent loci varied from 0.00 to 28.31 cM/Mb with an average of 4.10 cM/Mb (fig. 7B). No recombination was detected between loci spanning from 4.0 Mb of LG 19 to 7.0 Mb of LG 9, which resulted in a total length of 23.2 Mb. The recombinationally suppressed region reached 80.2% of LG 19 and 34.4% of LG 9, or 57.3% of LGs 19 and 9 combined. Furthermore, considerably low recombination rates (0.23–0.42 cM/Mb) were observed in the region between 7.0 Mb and 15.4 Mb in LG 9, resulting in a shorter linkage distance of this region in the male map (2.2 cM) than in the female map (49.9 cM). The recombination rate increased to 10.33 cM/Mb in the region between 15.4 Mb and 19.7 Mb and to 28.31 cM/Mb in the region between 19.7 Mb and 20.2 Mb in LG 9, although no recombination was found in the latter region in the female meiosis. Similarly, high recombination rates (16.90–21.69 cM/Mb) were observed in the first 4.0 Mb region of LG 19 (fig. 7B). In this region, the linkage distance in the male map was considerably longer (84.4 cM) than that in the female map (18.4 cM).
The patterns of differentiation between the sex chromosomes were further assessed by the level of genetic differentiation as measured by DEST (fig. 7B). In LG 19, significant genetic differentiation was observed in all loci except for the three loci located in the first 1.6 Mb region (fig. 7B). The DEST was relatively constant (0.35–0.45) across the region between 3.2 Mb and 7.0 Mb. Although considerably high DEST values (0.73–1.00) were observed for some loci, the DEST varied from 0.07 to 1.00 in the region between 7.4 Mb and 15.9 Mb irrespective of chromosome location. In this region, the lowest DEST value (0.07) was found for the locus where the numbers of X and Y chromosomal alleles were remarkably high (29 and 11, respectively). In LG 9, the highest DEST value (0.55) was observed for the locus located proximally at the left end (0.05 Mb) of this LG (fig. 7B). The DEST decreased gradually with increasing distance from the end in the first 5.7 Mb region (fig. 7B). No significant differentiation was observed in the region between 5.7 Mb and 20.2 Mb.
Discussion
The results provide an overview of the genetic differentiation and evolutionary dynamics of the ancestral and neo-sex chromosomes of threespine sticklebacks. Contrasting levels of differentiation were uncovered between the ancestral and neo-sex chromosomes: Although the ancestral sex chromosomes were highly and extensively differentiated in all divergent lineages, genetic differentiation was detected only in a small region of the neo-sex chromosomes. The integrative analyses of genetic differentiation and recombination restriction provided a detailed picture of the process of neo-sex chromosome differentiation. In what follows, we discuss these and related issues in the context of theoretical and empirical knowledge regarding the processes of sex chromosome differentiation and evolution.
Evolutionary Pattern of Ancestral Sex Chromosomes
Stickleback fishes have experienced the rapid turnover of sex determination and sex chromosome systems (Ross et al. 2009). As such, their sex chromosomes are believed to be at an early stage of evolution. However, our results demonstrated that the ancestral sex chromosomes of threespine sticklebacks are highly differentiated from each other throughout most of their length in all divergent lineages, as shown by the chromosome-wide distributions of loci exhibiting sex-specific allelic patterns. Had the sex chromosomes evolved rapidly within the last 2 My, one would expect to observe different allelic patterns at several loci among the divergent lineages. However, our results revealed that allelic patterns at many loci were identical among these lineages. In addition, a putative pseudoautosomal region was confined in the same chromosomal region in all lineages. Similarly, several loci with male-specific null alleles were commonly found in a large chromosomal region in which a large deletion has been identified in the Y chromosome of the Pacific lineage (Ross and Peichel 2008). Thus, our results suggest that the contemporary genetic differentiation and organizations of the ancestral sex chromosomes have been formed mainly before the divergence of the Japan Sea and other lineages. Furthermore, the phylogenetic analyses based on X and Y chromosomal alleles support the hypothetical scenario that the ancestral sex chromosomes have extensively diverged before the split of these lineages. A similar divergence pattern was observed in short fragment sequences around the putative sex-determining locus (Peichel et al. 2004), although this could be a consequence of a tight physical linkage between sequenced sites and the sex-determining locus. The results from our chromosome-wide survey demonstrate a consistent divergence pattern across the ancestral sex chromosomes, implying that the genetic differentiation detected throughout these chromosomes has evolved earlier than 2 Ma.
Although chromosomal organizations are apparently highly conserved in the ancestral sex chromosomes among the divergent lineages, different sex-specific allelic patterns were identified in some of the loci located in the central chromosomal region. All these different patterns were based on the difference in Y chromosomal amplification between the Japan Sea and other lineages. Because Y chromosomal null alleles were not biased toward a particular lineage, it is unlikely that these differences resulted from the fact that all primers were designed based on the genomic sequences of Pacific lineage individuals. Molecular cytogenetic analyses with the Pacific lineage suggested possible multiple inversions in the same region of the Y chromosome (Ross and Peichel 2008). Therefore, ancestral Y chromosome rearrangements might have occurred differently between the Japan Sea lineage and other lineages in this genomic region. Alternatively, degeneration of the Y chromosome might have occurred differently in this region. Furthermore, it is also possible that the rearrangements and degeneration of the ancestral Y chromosome in the Japan Sea lineage have been affected by the formation of neo-sex chromosomes. Our results also indicated varying levels of genetic differentiation between the ancestral sex chromosomes in the region where multiple inversions were identified in the Y chromosome (Ross and Peichel 2008). The different levels of differentiation might have resulted from the Y chromosome rearrangements, although other factors such as different mutation rates or evolutionary histories of the loci cannot be ruled out. Further comparative analyses of these divergent lineages with the Y chromosome sequences would provide insight into what kinds of rearrangements and degeneration have evolved before and after the split of these lineages.
As commonly observed in organisms with highly differentiated sex chromosomes (Bergero and Charlesworth 2009; Gschwend et al. 2012), recombination has been extensively suppressed between the ancestral sex chromosomes of the Japan Sea lineage. Our results demonstrated that recombination frequency during male meiosis is not only reduced in the sex-determining region but is also increased in the pseudoautosomal region of these chromosomes. Increased recombination in the heterogametic sex relative to the homogametic sex in a pseudoautosomal region has been reported in a limited number of organisms, such as the human and mouse (Ellis and Goodfellow 1989; Rappold 1993; Otto et al. 2011). Recombination frequency in a pseudoautosomal region is estimated to be 10-fold greater in males than females in the human and 7-fold greater in the mouse (Rouyer et al. 1986; Soriano et al. 1987). In fish, a similar result was observed in the Japanese medaka (Kondo et al. 2001). A possible explanation for this phenomenon is that at least one crossover is required for the proper segregation of the sex chromosomes during meiosis in many species, and consequently, recombination occurs exclusively in a small pseudoautosomal region in the heterogametic sex (Rouyer et al. 1986; Soriano et al. 1987). Our study showed that recombination frequency in the pseudoautosomal region is 4.6 times higher in male than in female meiosis in the threespine sticklebacks of the Japan Sea lineage. In contrast, the ratio was estimated to be 1.3 times in the males than females of the Pacific lineage (Otto et al. 2011). The considerable difference observed between these lineages could be due to the extensive recombination restriction across the ancestral and neo-sex chromosomes caused by the formation of a neo-sex chromosome system in the Japan Sea lineage.
Genetic Differentiation of Neo-sex Chromosomes
Clearly distinct levels and patterns of genetic differentiation were uncovered between the ancestral and neo-sex chromosomes of the Japan Sea lineage. In contrast to the ancestral sex chromosomes, loci with sex-specific allelic patterns were observed only in a small region at one end of the neo-sex chromosomes. Furthermore, no male-specific null alleles were detected at any loci, offering no indications of an identifiable inversion or deletion. In the region where male-specific alleles were identified, the level of genetic differentiation was highest at the end of the chromosomes and decreased with increasing distance from the chromosome end. Thus, our results suggest that genetic differentiation was initiated at the fusion point in the neo-sex chromosomes followed by gradual spread to neighboring regions. The analyses of recombination frequency further provided a detailed picture of the processes of recombination restriction and differentiation in the neo-sex chromosomes. For instance, it is evident that recombination restriction has extensively and continuously evolved in a large region spanning the ancestral and the neo-sex chromosomes, implying that the neo-Y chromosome of this region cosegregates with the sex-determining locus. In the neo-sex chromosomes, recombination is suppressed over a larger chromosomal region (7.0 Mb) than the region where genetic differentiation is observed (3.2 Mb). In addition, recombination is apparently reduced at a distance as far as 15.4 Mb. Because differentiation of sex chromosomes occurs after recombination suppression (Bengtsson and Goodfellow 1987), it appears that genetic differentiation of the neo-sex chromosomes has gradually spread from the end where the neo-Y chromosome has fused to the ancestral Y, following the extension of recombination suppression. Thus, our results suggest that the progression of differentiation between the neo-sex chromosomes of the threespine stickleback is gradual, rather than a consequence of large inversions or other chromosomal rearrangements.
A recent study of birds found that neo-sex chromosomes were formed in Sylvioidea species as a result of the fusion of a part of an autosome to the ancestral sex chromosomes (Pala, Naurin, et al. 2012). Similar to our results, differentiation patterns of the neo-sex chromosomes suggested that suppression of recombination has spread from the distal region closely proximate to the ancestral sex chromosomes (Pala, Hasselquist, et al. 2012). These observations are in line with the theoretical expectation that recombination between neo-sex chromosomes ceases due to translocation of autosomal elements into nonrecombining ancestral sex chromosomes, emphasizing the importance of physical linkage for the initial differentiation of neo-sex chromosomes (Rice 1987; Davisson and Akeson 1993; Vieira et al. 2003; Charlesworth and Mank 2010). Furthermore, selection that favors linkage between the sex-determining locus and sexually antagonistic genes might have promoted a reduction of recombination between the neo-sex chromosomes (Rice 1987; Bergero and Charlesworth 2009). Interestingly, it has been shown that the newly formed neo-sex chromosomes of the threespine stickleback are involved in male courtship behavior, which might be subject to sexually antagonistic selection (Kitano et al. 2009). However, a genomic region determining this trait is located on the opposite side of the chromosomal region to where differentiation and recombination suppression have been established. In addition, there is little evidence for linkage disequilibrium between sex-linked loci in the ancestral sex chromosomes and the genomic region determining courtship behavior in the neo-sex chromosomes. Thus, it is unlikely that restriction of recombination between the neo-sex chromosomes was induced by genes responsible for this trait.
The evolutionary dynamics of neo-sex chromosomes have been intensively studied in Drosophila as a model of sex chromosome evolution (Kaiser and Bachtrog 2010). Although the neo-sex chromosomes of Drosophila miranda were formed approximately 1 Ma due to a fusion between the ancestral Y chromosome and an autosome (Bachtrog and Charlesworth 2002), the entire region of these chromosomes has ceased recombining simultaneously after the rearrangement event, leading to a rapid differentiation (Bachtrog et al. 2008). Approximately half of the genes on the neo-Y chromosome have lost function due to the degeneration of this chromosome (Bachtrog et al. 2008). Moreover, dosage compensation has partially evolved in the neo-X chromosome (Steinemann M and Steinemann S 1998; Bachtrog 2006b). It has also been demonstrated that deleterious mutations have already accumulated in the neo-Y chromosome of D. albomicans, which was formed only 0.12 Ma (Zhou et al. 2012). Based on the assumption that the fusion between the ancestral Y and the neo-Y chromosomes had a role in the divergence of ancestral threespine sticklebacks of the Japan Sea and Pacific lineages (Kitano et al. 2009), a reduction of recombination between the neo-sex chromosomes could have initiated approximately 2 Ma. In contrast to Drosophila with achiasmatic meiotic systems, the progression patterns of recombination restriction and differentiation are apparently gradual and continuous in the neo-sex chromosomes of the threespine stickleback. It has been also suggested that the levels of differentiation and degeneration in ancestral sex chromosomes might affect the process of neo-sex chromosome differentiation (Charlesworth et al. 2005; Bachtrog 2011). In fact, the ancestral sex chromosomes of Drosophila are likely to be much older than those of the threespine stickleback, in which dosage compensation has not evolved yet (Leder et al. 2010). However, despite the highly evolved ancestral sex chromosomes of birds, the neo-sex chromosomes of the Sylvioidea appear to have been recombining over a considerable period of evolutionary time after the fusion event, exhibiting a gradual reduction of recombination as in the threespine stickleback (Pala, Hasselquist, et al. 2012). These studies highlight a variety of factors potentially influencing neo-sex chromosome evolution. However, because such empirical studies are still scarce, more work is required for a better understanding of the evolutionary dynamics of neo-sex chromosomes and molecular mechanisms underlying recombination suppression and differentiation.
Microsatellite analyses provided a clear picture of the differentiation state of evolutionarily young neo-sex chromosomes. In addition, phylogenetic analyses based on these loci verified the evolutionary origin and divergence of these chromosomes. The great advantage of microsatellites lies in their high mutation rate, as estimated to be approximately 10−4 in fish (Estoup and Angers 1998; Shimoda et al. 1999; Lippe et al. 2006). This rate is considerably higher than that of single-nucleotide polymorphisms, which is considered to be 10−8–10−10 (Baer et al. 2007). Despite a progressive reduction of recombination in up to approximately 2 My of evolutionary time, allelic reduction and differentiation could be apparent at microsatellite loci owing to their high sensitivity to population bottlenecks and genetic drift, which are expected to increase in nonrecombining sex chromosomes due to their smaller effective population sizes when compared with autosomes (Ellegren 2009). In addition, new mutations could arise at microsatellite loci even in such a short evolutionary time scale, which would increase the chance to minimize the background signatures of ancestrally shared polymorphisms (Schlötterer 2000). Our study identified a gap between the regions showing genetic differentiation and recombination restriction in the neo-sex chromosomes. This gap could result from a time lag until certain alleles or new mutations accumulate on different neo-sex chromosomes after recombination cessation. In contrast to microsatellites, less polymorphic markers with lower mutation rates are likely to be less sensitive in detecting genetic differentiation in recently evolved sex chromosomes.
Conclusions
Our comparative analyses of different threespine stickleback lineages revealed genetic transitions in the ancestral and neo-sex chromosomes in a short evolutionary time period. The results further suggest that the processes of reduced recombination and genetic differentiation between the neo-sex chromosomes are gradual, rather than a consequence of large inversions or other chromosomal rearrangements. Thus, our study empirically supports the theoretical model that recombination suppression and differentiation proceed in a gradual manner in the very early stage of sex chromosome evolution. The findings also suggest that recombination restriction in the neo-sex chromosomes was induced by the physical linkage between the ancestral and neo-Y chromosomes. In addition, although the ancestral sex chromosomes are highly differentiated and evolutionarily stable, recombination in the pseudoautosomal region is likely accelerated due to extensive and continuous recombination suppression across the ancestral and neo-sex chromosomes as a consequence of the new formation of a neo-sex chromosome system. Thus, our results emphasize the potential importance of interaction between the ancestral and neo-sex chromosomes in their evolutionary context. Based on our study, the sex chromosome system of the Japan Sea lineage promises to serve as an important model for further understanding of the molecular processes of sex chromosome differentiation and evolution.
Materials and Methods
Samples
Threespine sticklebacks were collected from nine globally distributed populations using minnow traps and nets (fig. 1 and supplementary table S1, Supplementary Material online). These populations were classified into the Pacific (P-SHI, P-TOW, and P-WES), Atlantic (A-HEL, A-KIN, A-MYV, and A-MAI), and Japan Sea lineages (J-BIW and J-MAS), based on their distribution and morphological characteristics (Haglund et al. 1992; Higuchi and Goto 1996; Higuchi et al. 1996). For each population, 46–54 adult fish were used for genetic analyses (supplementary table S1, Supplementary Material online). Phenotypic sex was determined by secondary sex characteristics and visual inspections of the gonads. Genetic differentiation of sex chromosomes were intensively analyzed using three populations (P-SHI, A-HEL, and J-BIW) representing the three divergent lineages with 63 microsatellite loci. Genetic analyses of the remaining six populations were conducted using the 23 loci where male-specific alleles were detected for at least one population in the initial survey (see Results for details). This enabled comparative analyses on the amplification and differentiation of X and Y chromosomal alleles at the 23 loci across the nine populations, as well as phylogenetic analyses based on X and Y chromosomal alleles at these loci. Genotyping data for 14 loci in A-HEL population were obtained from a previous study (Shikano et al. 2011).
For recombination and linkage analyses in the ancestral and neo-sex chromosomes of the Japan Sea lineage, three full-sib families were produced in J-BIW population by means of artificial crosses with wild parental fish according to Shimada, Shikano, Kuparinen, et al. (2011). These F1 fish were held in three 90 -l aquaria at 17 °C up to 400 days after hatching. In total, 197 individuals consisting of six parents and the191 F1 offspring (63 or 64 per family) were genotyped for 36 loci in which no male-specific null alleles were identified in this population (see Results).
Genotyping
Tissue samples were digested with proteinase K, and total genomic DNA was extracted using a silica-based purification method (Elphinstone et al. 2003). A total of 63 microsatellite loci, comprising 36 loci on LG 19 and 27 on LG9, were selected to span widely these chromosomes based on their physical locations in the reference genome sequence of the threespine stickleback (supplementary table S2, Supplementary Material online). Of these, 38 loci were chosen from previous studies (Peichel et al. 2001; Mäkinen et al. 2008; Shikano et al. 2011; Shimada, Shikano, Merilä, 2011). Novel primer pairs were developed for additional 25 loci with the genome sequence using WebSat (Martins et al. 2009). These primers were deposited in the National Center for Biotechnology Information Probe Database (PUIDs 16584713–16584737; supplementary table S2, Supplementary Material online). Polymerase chain reaction (PCR) amplifications of microsatellite loci were conducted using the Qiagen Multiplex PCR Kit (Qiagen) in a reaction volume of 10 µl consisting of approximately 20 ng of template DNA, 1× Multiplex PCR Master Mix, 0.5× Q-Solution, and 2 pmol of each primer. Thermocycling conditions were as follows: initial denaturation at 95 °C for 15 min, followed by 30 cycles of 30 s at 94 °C, 90 s at 53 °C, and 60 s at 72 °C and a final extension at 60 °C for 5 min. Amplified fragments were visualized with a MegaBACE 1000 automated sequencer (Amersham Biosciences) using ET-ROX 550 size standard (Amersham Biosciences) and analyzed using Fragment Profiler 1.2 (Amersham Biosciences). All alleles were scored by H.M.N. and double checked by T.S.
Data Analyses
The number of alleles, observed and expected heterozygosities, and FIS were calculated using FSTAT 2.9.3 (Goudet 1995). The significance of FIS was determined by 10,000 permutations. These parameters were estimated for each population, as well as for respective sexes to investigate amplification of X and Y chromosomal alleles and differentiation between these alleles. Linkage disequilibrium between all pairs of loci was tested in each population using Genepop 4.0 (Rousset 2008). Genetic differentiation between sex chromosomes was evaluated based on FST between females and males, as well as the presence of male-specific alleles. FST values and their significance were determined using Genepop with Markov chain parameters of 10,000 dememorization steps, 100 batches, and 5,000 iterations per patch. Male-specific alleles were identified by comparing allele distributions and genotypic frequencies between males and females using GenAlEx 6.5 (Peakall and Smouse 2012). In theory, FIS has a tendency to be negative in heterogametic males but not in homogametic females when Y-specific alleles are present. In the regions where Y chromosome degeneration, deletion, or inversion has occurred, no heterozygous individuals can be observed in males due to no amplification of Y chromosomal alleles. Accordingly, observed heterozygosity and FIS are expected to be largely different between sexes in these regions, exhibiting significantly positive FIS in males but not in females. In contrast, significantly positive FIS values can be found independently of sex when null alleles are commonly present in both X and Y chromosomes. Because such patterns were identified at Gasm14 and GR1 in P-SHI, at Gasm10 and Gasm30 in A-HEL, and at Gasm28 and Gasm29 in J-BIW (supplementary table S3, Supplementary Material online), these loci were removed from the analyses of sex chromosome differentiation. In addition, because no amplification or polymorphism was detected for Gasm3, Gasm17, PKMa, Gasm11, Gasm7, and Gasm22 in J-BIW, for Gasm16 and Gasm26 in P-TOW, and for HSP90B and Gasm26 in A-MYV (supplementary tables S3 and S8, Supplementary Material online), these loci were not included in the analyses. Standard Bonferroni corrections were applied for all multiple comparisons.
To address whether the ancestral sex chromosomes have genetically differentiated from each other before or after the divergence of the three lineages, phylogenetic relationships were assessed in terms of sex chromosomes and populations using Cavalli-Sforza–Edwards distances (DCE; Cavalli-Sforza and Edwards 1967), which do not assume constant population sizes or constant mutation rates across loci. This analysis was conducted based on the X and Y chromosomal alleles identified in males using the loci where male-specific alleles were detected. Phylogenetic relationships were constructed with the NJ method using Populations 1.2.31 (Langella 1999). Confidence in tree nodes was determined with 1,000 bootstraps across loci. Phylogenetic relationships were also investigated for each locus to assess whether heterogeneous differentiation patterns are observed across the ancestral sex chromosomes. In addition, phylogenetic analyses were conducted for neo-sex chromosomes using the neo-X and neo-Y chromosomal alleles identified in males. Because no sex-specific alleles were expected to be observed in the Pacific and Atlantic lineages due to the absence of the neo-sex chromosome system, male data of these populations were used for the analyses.
Recombination and linkage analyses in the Japan Sea lineage were conducted for female and male meioses using JoinMap 4.1 (Stam 1993; Van Ooijen 2006). In each family, pairwise recombination frequencies were calculated based on a threshold range from 0.50 to 0.00 with a step value of −0.05. Linkage between loci was detected based on these estimates. In all the three full-sib families used for the analyses, two LGs were identified in the female meiosis and one in the male meiosis. Locus orders and distances in consensus linkage maps were determined by the mean recombination frequencies and combined LOD scores of three family data using the regression mapping approach with a LOD threshold of 3.0 and goodness-of-fit jump threshold of 3.0. In the consensus analyses, significant linkage was detected for 26 loci in the female meiosis and 20 loci in the male meiosis. The Kosambi mapping function was used to convert recombination frequencies to map distances in cM. Recombination rates were given as cM/Mb based on the genetic and physical distances between adjacent loci. Recombination frequencies in a pseudoautosomal region were calculated for female and male meioses based on map distances following Otto et al. (2011).
For comprehensive analyses of sex chromosome differentiation in the Japan Sea lineage, the degree and patterns of genetic differentiation between sex chromosomes were assessed based on X and Y chromosome haplotypes. The pseudo-Bayesian ELB algorithm as implemented in Arlequin 3.5 (Excoffier and Lischer 2010) was used to determine the most probable haplotype constitution (Excoffier et al. 2003). The analysis was conducted for LGs 19 and 9 separately under the following settings: Dirichlet prior alpha value, 0.01; epsilon value, 0.2; heterozygote site influence zone, 5; gamma value, 0.01; sampling interval, 500; no. of samples, 2,000; burn-in steps, 100,000; and recombination steps, 0%. Loci with male-specific null alleles were removed from the analysis. Two loci, Stn235 and Stn190, on LG 19 were not included in the haplotype estimation because male-specific alleles were erroneously assigned to X chromosome alleles due to no polymorphism in females. The X and Y haplotypes of each individual were determined based on male-specific alleles. The level of genetic differentiation between X and Y chromosomes was evaluated using the actual differentiation DEST proposed by Jost (2008), which is independent of heterozygosity, and is thus suitable for highly polymorphic markers such as microsatellites (Jost 2008; Gerlach et al. 2010). The analysis was performed using SMOGD (Crawford 2010) with 1,000 bootstraps to estimate the 95% confidence interval of each locus.
As the genome sequence of the Y chromosome is not available in the threespine stickleback, the physical locations of marker loci were determined based on the reference genome sequence of a female individual in the Pacific lineage (supplementary table S2, Supplementary Material online). The orientation of six loci in the supercontig 3 (3.8–20.2 Mb) of LG 19 in our genetic linkage map was inverted when compared with the reference genome sequence (see Results) due to a possible error in the genome sequence (Ross and Peichel 2008), as observed for the loci of this supercontig in a linkage map of the Pacific lineage (Peichel et al. 2004). Therefore, marker locations on LG 19 were determined by reversing the supercontig 3 sequence (supplementary table S2, Supplementary Material online), according to Ross and Peichel (2008). Similarly, the orientation of 10 loci in the supercontig 8 (4.3–17.9 Mb) of LG 9 was consistently reversed when compared with the reference genome sequence (see Results). Because the order of loci in this region is also inverted in a linkage map of the Pacific lineage (Peichel et al. 2001), this could be due to an error in the reference genome sequence rather than a lineage-specific chromosomal rearrangement. Therefore, the same correction was implemented for marker locations on this supercontig (supplementary table S2, Supplementary Material online).
Supplementary Material
Supplementary tables S1–S9 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Charles Baube, Akira Goto, Frank von Hippel, Takefumi Kitamura, Yuichiro Meguro, Antoine Millet, Katja Räsänen, Garrett Staines, Mike Webster, and James Willacker for help in obtaining samples. They also thank Jacquelin DeFaveri for correcting the English. This work was supported by the Academy of Finland to J.M., the Otto A. Malm Foundation to H.M.N., and the Finnish Graduate School in Population Genetics to H.M.N.
References
- Andolfatto P, Depaulis F, Navarro A. Inversion polymorphisms and nucleotide variability in Drosophila. Genet Res. 2001;77:1–8. doi: 10.1017/s0016672301004955. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. A dynamic view of sex chromosome evolution. Curr Opin Genet Dev. 2006a;16:578–585. doi: 10.1016/j.gde.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. Expression profile of a degenerating neo-Y chromosome in Drosophila. Curr Biol. 2006b;16:1694–1699. doi: 10.1016/j.cub.2006.07.053. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. Plant sex chromosomes: a non-degenerated Y? Curr Biol. 2011;21:R685–R688. doi: 10.1016/j.cub.2011.08.027. [DOI] [PubMed] [Google Scholar]
- Bachtrog D, Charlesworth B. Reduced adaptation of a nonrecombining neo-Y chromosome. Nature. 2002;416:323–326. doi: 10.1038/416323a. [DOI] [PubMed] [Google Scholar]
- Bachtrog D, Hom E, Wong KM, Maside X, de Jong P. Genomic degradation of a young Y chromosome in Drosophila miranda. Genome Biol. 2008;9:R30. doi: 10.1186/gb-2008-9-2-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer CF, Miyamoto MM, Denver DR. Mutation rate variation in multicellular eukaryotes: causes and consequences. Nat Rev Genet. 2007;8:619–631. doi: 10.1038/nrg2158. [DOI] [PubMed] [Google Scholar]
- Bengtsson BO, Goodfellow PN. The effect of recombination between the X and Y chromosomes of mammals. Ann Hum Genet. 1987;51:57–64. doi: 10.1111/j.1469-1809.1987.tb00865.x. [DOI] [PubMed] [Google Scholar]
- Bergero R, Charlesworth D. The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol. 2009;24:94–102. doi: 10.1016/j.tree.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Bergero R, Charlesworth D, Filatov DA, Moore RC. Defining regions and rearrangements of the Silene latifolia Y chromosome. Genetics. 2008;178:2045–2053. doi: 10.1534/genetics.107.084566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R, Forrest A, Kamau E, Charlesworth D. Evolutionary strata on the X chromosomes of the dioecious plant Silene latifolia: evidence from new sex-linked genes. Genetics. 2007;175:1945–1954. doi: 10.1534/genetics.106.070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ. Evolution of sex determining mechanisms. Menlo Park (CA): Benjamin Cummings; 1983. [Google Scholar]
- Cavalli-Sforza LL, Edwards AW. Phylogenetic analysis: models and estimation procedures. Am J Hum Genet. 1967;19:233–257. [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. The evolution of chromosomal sex determination and dosage compensation. Curr Biol. 1996;6:149–162. doi: 10.1016/s0960-9822(02)00448-7. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. Sex determination: primitive Y chromosomes in fish. Curr Biol. 2004;14:R745–R747. doi: 10.1016/j.cub.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Hartl DL. Population-dynamics of the segregation distorter polymorphism of Drosophila melanogaster. Genetics. 1978;89:171–192. doi: 10.1093/genetics/89.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Mank J. The birds and the bees and the flowers and the trees: lessons from genetic mapping of sex determination in plants and animals. Genetics. 2010;186:9–31. doi: 10.1534/genetics.110.117697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioffi MB, Moreira-Filho O, Almeida-Toledo LF, Bertollo LAC. The contrasting role of heterochromatin in the differentiation of sex chromosomes: an overview from Neotropical fishes. J Fish Biol. 2012;80:2125–2139. doi: 10.1111/j.1095-8649.2012.03272.x. [DOI] [PubMed] [Google Scholar]
- Cnaani A, Lee BY, Zilberman N, et al. (15 co-authors) Genetics of sex determination in tilapiine species. Sex Dev. 2008;2:43–54. doi: 10.1159/000117718. [DOI] [PubMed] [Google Scholar]
- Crawford NG. SMOGD: software for the measurement of genetic diversity. Mol Ecol Resour. 2010;10:556–557. doi: 10.1111/j.1755-0998.2009.02801.x. [DOI] [PubMed] [Google Scholar]
- Davisson MT, Akeson EC. Recombination suppression by heterozygous Robertsonian fusions in the mouse. Genetics. 1993;133:649–667. doi: 10.1093/genetics/133.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deagle BE, Reimchen TE, Levin DB. Origins of endemic stickleback from the Queen Charlotte Islands: mitochondrial and morphological evidence. Can J Zool. 1996;74:1045–1056. [Google Scholar]
- Devlin RH, Nagahama Y. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture. 2002;208:191–364. [Google Scholar]
- Ellegren H. The different levels of genetic diversity in sex chromosomes and autosomes. Trends Genet. 2009;25:278–284. doi: 10.1016/j.tig.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Ellegren H. Sex-chromosome evolution: recent progress and the influence of male and female heterogamety. Nat Rev Genet. 2011;12:157–166. doi: 10.1038/nrg2948. [DOI] [PubMed] [Google Scholar]
- Ellis N, Goodfellow PN. The mammalian pseudoautosomal region. Trends Genet. 1989;5:406–410. doi: 10.1016/0168-9525(89)90199-6. [DOI] [PubMed] [Google Scholar]
- Elphinstone MS, Hinten GN, Anderson MJ, Nock CJ. An inexpensive and high-throughput procedure to extract and purify total genomic DNA for population studies. Mol Ecol Notes. 2003;3:317–320. [Google Scholar]
- Estoup A, Angers B. Microsatellites and minisatellites for molecular ecology: theoretical and empirical considerations. In: Carvlho GR, editor. Advances in molecular ecology. NATO Science Series. Amsterdam (The Netherlands): IOS Press; 1998. pp. 55–86. [Google Scholar]
- Excoffier L, Laval G, Balding D. Gametic phase estimation over large genomic regions using an adaptive window approach. Hum Genomics. 2003;1:7–19. doi: 10.1186/1479-7364-1-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Fisher RA. The evolution of dominance. Biol Rev. 1931;6:345–368. [Google Scholar]
- Gerlach G, Jueterbock A, Kraemer P, Deppermann J, Harmand P. Calculations of population differentiation based on GST and D: forget GST but not all of statistics! Mol Ecol. 2010;19:3845–3852. doi: 10.1111/j.1365-294X.2010.04784.x. [DOI] [PubMed] [Google Scholar]
- Goudet J. FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered. 1995;86:485–486. [Google Scholar]
- Gschwend AR, Weingartner LA, Moore RC, Ming R. The sex-specific region of sex chromosomes in animals and plants. Chromosome Res. 2012;20:57–69. doi: 10.1007/s10577-011-9255-y. [DOI] [PubMed] [Google Scholar]
- Haglund TR, Buth DG, Lawson R. Allozyme variation and phylogenetic relationships of Asian, North American, and European populations of the threespine stickleback. Copeia. 1992;1992:432–443. [Google Scholar]
- Handley L-JL, Ceplitis H, Ellegren H. Evolutionary strata on the chicken Z chromosome: implications for sex chromosome evolution. Genetics. 2004;167:367–376. doi: 10.1534/genetics.167.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Goto A. Genetic evidence supporting the existence of two distinct species in the genus Gasterosteus around Japan. Environ Biol Fishes. 1996;47:1–16. [Google Scholar]
- Higuchi M, Goto A, Yamazaki F. Genetic structure of threespine stickleback, Gasterosteus aculeatus, in Lake Harutori, Japan, with reference to coexisting anadromous and freshwater forms. Ichthyol Res. 1996;43:349–358. [Google Scholar]
- Janousek B, Mrackova M. Sex chromosomes and sex determination pathway dynamics in plant and animal models. Biol J Linn Soc. 2010;100:737–752. [Google Scholar]
- Jobling MA, Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet. 2003;4:598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- Jost L. GST and its relatives do not measure differentiation. Mol Ecol. 2008;17:4015–4026. doi: 10.1111/j.1365-294x.2008.03887.x. [DOI] [PubMed] [Google Scholar]
- Kaiser VB, Bachtrog D. Evolution of sex chromosomes in insects. Annu Rev Genet. 2010;44:91–112. doi: 10.1146/annurev-genet-102209-163600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura Y, Iwase M, Satta Y. Evolution of genomic structures on Mammalian sex chromosomes. Curr Genomics. 2012;13:115–123. doi: 10.2174/138920212799860625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano J, Peichel CL. Turnover of sex chromosomes and speciation in fishes. Environ Biol Fishes. 2012;94:549–558. doi: 10.1007/s10641-011-9853-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano J, Ross JA, Mori S, et al. (12 co-authors) A role for a neo-sex chromosome in stickleback speciation. Nature. 2009;461:1079–1083. doi: 10.1038/nature08441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Nagao E, Mitani H, Shima A. Differences in recombination frequencies during female and male meioses of the sex chromosomes of the medaka, Oryzias latipes. Genet Res. 2001;78:23–30. doi: 10.1017/s0016672301005109. [DOI] [PubMed] [Google Scholar]
- Kondo M, Nanda I, Schmid M, Schartl M. Sex determination and sex chromosome evolution: insights from medaka. Sex Dev. 2009;3:88–98. doi: 10.1159/000223074. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Langella O. Populations 1.2.28: a population genetic software. 1999 CNRS UPR9034. [cited 2013 March 8]. Available from: http://bioinformatics.org/∼tryphon/populations/ [Google Scholar]
- Leder EH, Cano JM, Leinonen T, O'Hara R, Primmer CR, Merilä J. Female-biased expression on the X chromosome as a key step in sex chromosome evolution in threespine sticklebacks. Mol Biol Evol. 2010;27:1495–1503. doi: 10.1093/molbev/msq031. [DOI] [PubMed] [Google Scholar]
- Lemaitre C, Braga MDV, Gautier C, Sagot MF, Tannier E, Marais GAB. Footprints of inversions at present and past pseudoautosomal boundaries in human sex chromosomes. Genome Biol Evol. 2009;1:56–66. doi: 10.1093/gbe/evp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippe C, Dumont P, Bernatchez L. High genetic diversity and no inbreeding in the endangered copper redhorse, Moxostoma hubbsi (Catostomidae, Pisces): the positive sides of a long generation time. Mol Ecol. 2006;15:1769–1780. doi: 10.1111/j.1365-294X.2006.02902.x. [DOI] [PubMed] [Google Scholar]
- Mäkinen HS, Cano JM, Merilä J. Identifying footprints of directional and balancing selection in marine and freshwater three-spined stickleback (Gasterosteus aculeatus) populations. Mol Ecol. 2008;17:3565–3582. doi: 10.1111/j.1365-294X.2008.03714.x. [DOI] [PubMed] [Google Scholar]
- Mank JE, Avise JC. Evolutionary diversity and turn-over of sex determination in teleost fishes. Sex Dev. 2009;3:60–67. doi: 10.1159/000223071. [DOI] [PubMed] [Google Scholar]
- Mank JE, Promislow DE, Avise JC. Evolution of alternative sex-determining mechanisms in teleost fishes. Biol J Linn Soc. 2006;87:83–93. [Google Scholar]
- Martins WS, Lucas DC, Neves KF, Bertioli DJ. WebSat—a web software for microsatellite marker development. Bioinformation. 2009;3:282–283. doi: 10.6026/97320630003282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K, Ellegren H. The chicken (Gallus gallus) Z chromosome contains at least three nonlinear evolutionary strata. Genetics. 2008;180:1131–1136. doi: 10.1534/genetics.108.090324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, Betrán E, Barbadilla A, Ruiz A. Recombination and gene flux caused by gene conversion and crossing over in inversion heterokaryotypes. Genetics. 1997;146:695–709. doi: 10.1093/genetics/146.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas M, Marais G, Hykelova V, Janousek B, Laporte V, Vyskot B, Mouchiroud D, Negrutiu I, Charlesworth D, Monéger F. A gradual process of recombination restriction in the evolutionary history of the sex chromosomes in dioecious plants. PLoS Biol. 2005;3:e4. doi: 10.1371/journal.pbio.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution of sex chromosomes in mammals. Annu Rev Genet. 1969;3:495–524. [Google Scholar]
- O'Reilly P, Reimchen TE, Beech R, Strobeck C. Mitochondrial DNA in Gasterosteus and Pleistocene glacial refugium on the Queen Charlotte Islands, British Columbia. Evolution. 1993;47:678–684. doi: 10.1111/j.1558-5646.1993.tb02122.x. [DOI] [PubMed] [Google Scholar]
- Ortí G, Bell MA, Reimchen TE, Meyer A. Global survey of mitochondrial-DNA sequences in the threespine stickleback: evidence for recent migrations. Evolution. 1994;48:608–622. doi: 10.1111/j.1558-5646.1994.tb01348.x. [DOI] [PubMed] [Google Scholar]
- Otto SP, Pannell JR, Peichel CL, Ashman T-L, Charlesworth D, Chippindale AK, Delph LF, Guerrero RF, Scarpino SV, McAllister BF. About PAR: the distinct evolutionary dynamics of the pseudoautosomal region. Trends Genet. 2011;27:358–367. doi: 10.1016/j.tig.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Pala I, Hasselquist D, Bensch S, Hansson B. Patterns of molecular evolution of an avian neo-sex chromosome. Mol Biol Evol. 2012;29:3741–3754. doi: 10.1093/molbev/mss177. [DOI] [PubMed] [Google Scholar]
- Pala I, Naurin S, Stervander M, Hasselquist D, Bensch S, Hansson B. Evidence of a neo-sex chromosome in birds. Heredity. 2012;108:264–272. doi: 10.1038/hdy.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearks Wilkerson AJ, Raudsepp T, Graves T, Albracht D, Warren W, Chowdhary BP, Skow LC, Murphy WJ. Gene discovery and comparative analysis of X-degenerate genes from the domestic cat Y chromosome. Genomics. 2008;92:329–338. doi: 10.1016/j.ygeno.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Peichel CL, Nereng K, Ohgi KA, Cole BLE, Colosimo PF, Buerkle CA, Schluter D, Kingsley DM. The genetic architecture of divergence between threespine stickleback species. Nature. 2001;414:901–905. doi: 10.1038/414901a. [DOI] [PubMed] [Google Scholar]
- Peichel CL, Ross JA, Matson CK, Dickson M, Grimwood J, Schmutz J, Myers RM, Mori S, Schluter D, Kingsley DM. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr Biol. 2004;14:1416–1424. doi: 10.1016/j.cub.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Rappold GA. The pseudoautosomal regions of the human sex chromosomes. Hum Genet. 1993;4:315–324. doi: 10.1007/BF01247327. [DOI] [PubMed] [Google Scholar]
- Rice WR. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution. 1987;41:911–914. doi: 10.1111/j.1558-5646.1987.tb05864.x. [DOI] [PubMed] [Google Scholar]
- Rice WR. Evolution of the Y sex chromosome in animals. BioScience. 1996;46:331–343. [Google Scholar]
- Ross JA, Peichel CL. Molecular cytogenetic evidence of rearrangements on the Y chromosome of threespine stickleback fish. Genetics. 2008;179:2173–2182. doi: 10.1534/genetics.108.088559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JA, Urton JR, Boland J, Shapiro MD, Peichel CL. Turnover of sex chromosomes in the stickleback fishes (Gasterosteidae) PLoS Genet. 2009;5:e1000391. doi: 10.1371/journal.pgen.1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross MT, Grafham CV, Coffey AK, et al. (283 co-authors) The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F. Genepop'007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resour. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Rouyer F, Simmler MC, Johnsson C, Vergnaud G, Cooke HJ, Weissenbach J. A gradient of sex linkage in the pseudoautosomal region of the human sex chromosomes. Nature. 1986;319:291–295. doi: 10.1038/319291a0. [DOI] [PubMed] [Google Scholar]
- Sandstedt SA, Tucker PK. Evolutionary strata on the mouse X chromosome correspond to strata on the human X chromosome. Genome Res. 2004;14:267–272. doi: 10.1101/gr.1796204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlötterer C. Microsatellite analysis indicates genetic differentiation of the neo-sex chromosomes in Drosophila americana americana. Heredity. 2000;85:610–616. doi: 10.1046/j.1365-2540.2000.00797.x. [DOI] [PubMed] [Google Scholar]
- Shikano T, Natri HM, Shimada Y, Merilä J. High degree of sex chromosome differentiation in stickleback fishes. BMC Genomics. 2011;12:474. doi: 10.1186/1471-2164-12-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Shikano T, Kuparinen A, Gonda A, Leinonen T, Merilä J. Quantitative genetics of body size and timing of maturation in two nine-spined stickleback (Pungitius pungitius) populations. PLoS One. 2011;6:e28859. doi: 10.1371/journal.pone.0028859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Shikano T, Merilä J. A high incidence of selection on physiologically important genes in the three-spined stickleback, Gasterosteus aculeatus. Mol Biol Evol. 2011;28:181–193. doi: 10.1093/molbev/msq181. [DOI] [PubMed] [Google Scholar]
- Shimoda N, Knapik EW, Ziniti J, Sim C, Yamada E, Kaplan S, Jackson D, deSauvage F, Jacob H, Fishman MC. Zebrafish genetic map with 2000 microsatellite markers. Genomics. 1999;58:219–232. doi: 10.1006/geno.1999.5824. [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx P, et al. (40 co-authors) The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Soriano P, Keitges EA, Schorderet DF, Harbers K, Gartler SM, Jaenisch R. High rate of recombination and double crossovers in the mouse pseudoautosomal region during male meiosis. Proc Natl Acad Sci U S A. 1987;84:7218–7220. doi: 10.1073/pnas.84.20.7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam P. Construction of integrated genetic linkage maps by means of a new computer package: JoinMap. Plant J. 1993;3:739–744. [Google Scholar]
- Steinemann M, Steinemann S. Enigma of Y chromosome degeneration: neo-Y and neo-X chromosomes of Drosophila miranda a model for sex chromosome evolution. Genetica. 1998;102/103:409–420. [PubMed] [Google Scholar]
- Stöck M, Horn A, Grossen C, et al. (14 co-authors) Ever-young sex chromosomes in European tree frogs. PLoS Biol. 2011;9:e1001062. doi: 10.1371/journal.pbio.1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehana Y, Demiyah D, Naruse K, Hamaguchi S, Sakaizumi M. Evolution of different Y chromosomes in two medaka species, Oryzias dancena and O. latipes. Genetics. 2007;175:1335–1340. doi: 10.1534/genetics.106.068247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Kivisild T. Use of Y chromosome and mitochondrial DNA population structure in tracing human migrations. Annu Rev Genet. 2007;41:539–564. doi: 10.1146/annurev.genet.41.110306.130407. [DOI] [PubMed] [Google Scholar]
- Van Laere AS, Coppieters W, Georges M. Characterization of the bovine pseudoautosomal boundary: documenting the evolutionary history of mammalian sex chromosomes. Genome Res. 2008;18:1884–1895. doi: 10.1101/gr.082487.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen J. JoinMap 4. Software for the calculation of genetic linkage maps in experimental populations. Wageningen (The Netherlands): Kyazma; 2006. [Google Scholar]
- Vieira CP, Coelho PA, Vieira J. Inferences on the evolutionary history of the Drosophila americana polymorphic X/4 fusion from patterns of polymorphism at the X-linked paralytic and elav genes. Genetics. 2003;164:1459–1469. doi: 10.1093/genetics/164.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff JN, Nanda I, Schmid M, Schartl M. Governing sex determination in fish: regulatory putsches and ephemeral dictators. Sex Dev. 2007;1:85–99. doi: 10.1159/000100030. [DOI] [PubMed] [Google Scholar]
- Wilson MA, Makova KD. Evolution and survival on eutherian sex chromosomes. PLoS Genet. 2009;5:e1000568. doi: 10.1371/journal.pgen.1000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer R, Kirsch S, Rappold GA, Schempp W. Evolutionary breakpoint analysis on Y chromosomes of higher primates provides insight into human Y evolution. Cytogenet Genome Res. 2005;108:204–210. doi: 10.1159/000080817. [DOI] [PubMed] [Google Scholar]
- Woram RA, Gharbi K, Sakamoto T, et al. (16 co-authors) Comparative genome analysis of the primary sex-determining locus in salmonid fishes. Genome Res. 2003;13:272–280. doi: 10.1101/gr.578503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Bachtrog D. Chromosome-wide gene silencing initiates Y degeneration in Drosophila. Curr Biol. 2012;22:522–525. doi: 10.1016/j.cub.2012.01.057. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Zhu H, Huang Q, et al. (16 co-authors) Deciphering neo-sex and B chromosome evolution by the draft genome of Drosophila albomicans. BMC Genomics. 2012;13:109. doi: 10.1186/1471-2164-13-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.