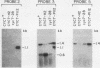
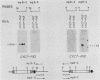
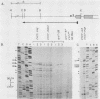
Abstract
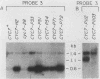
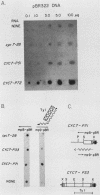
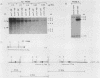
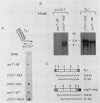
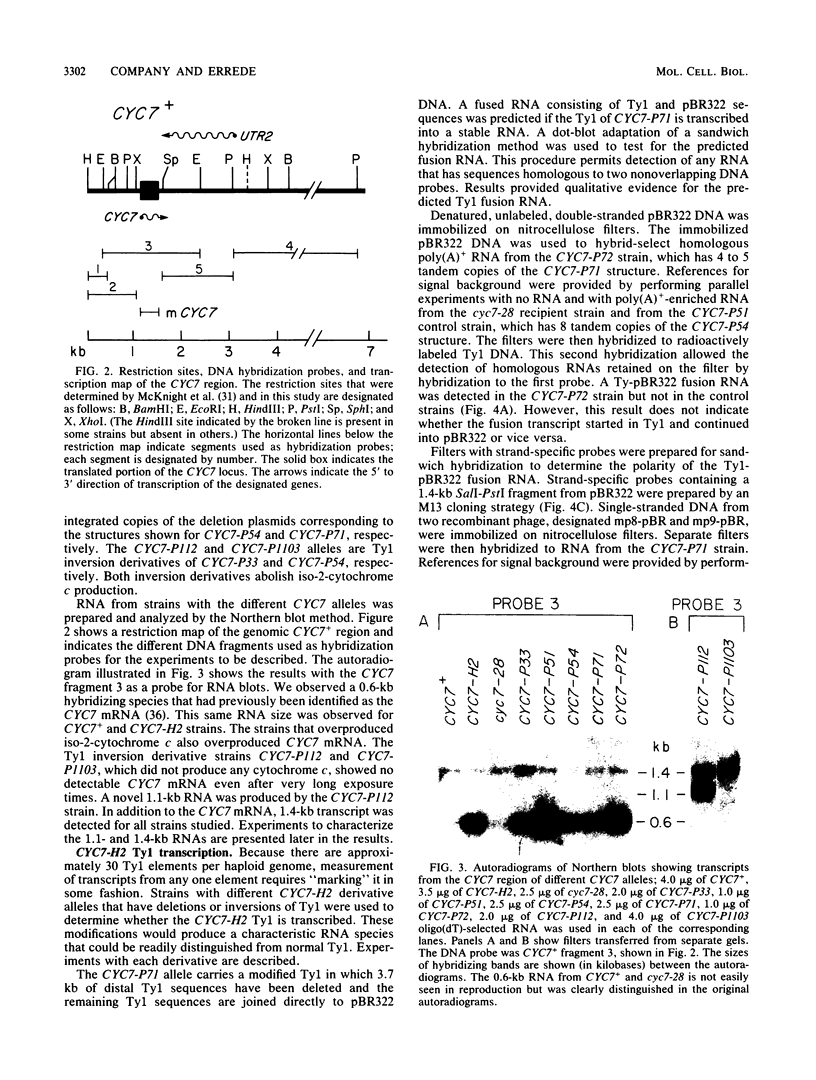
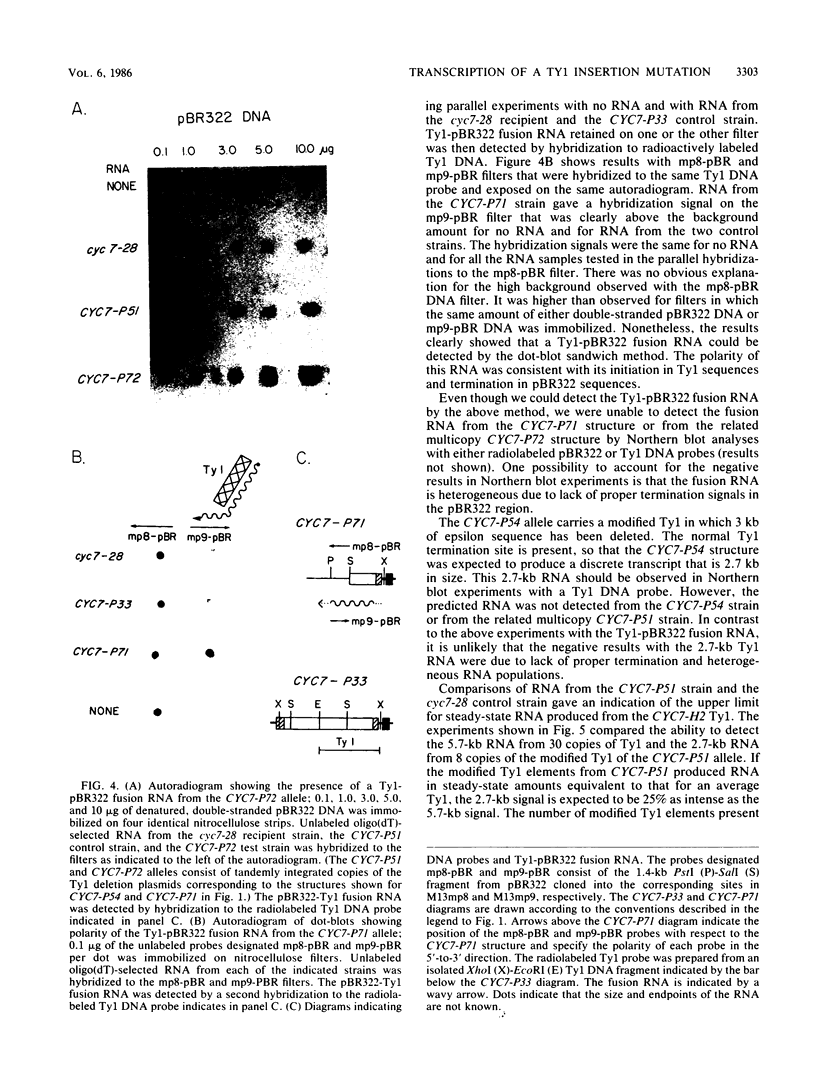
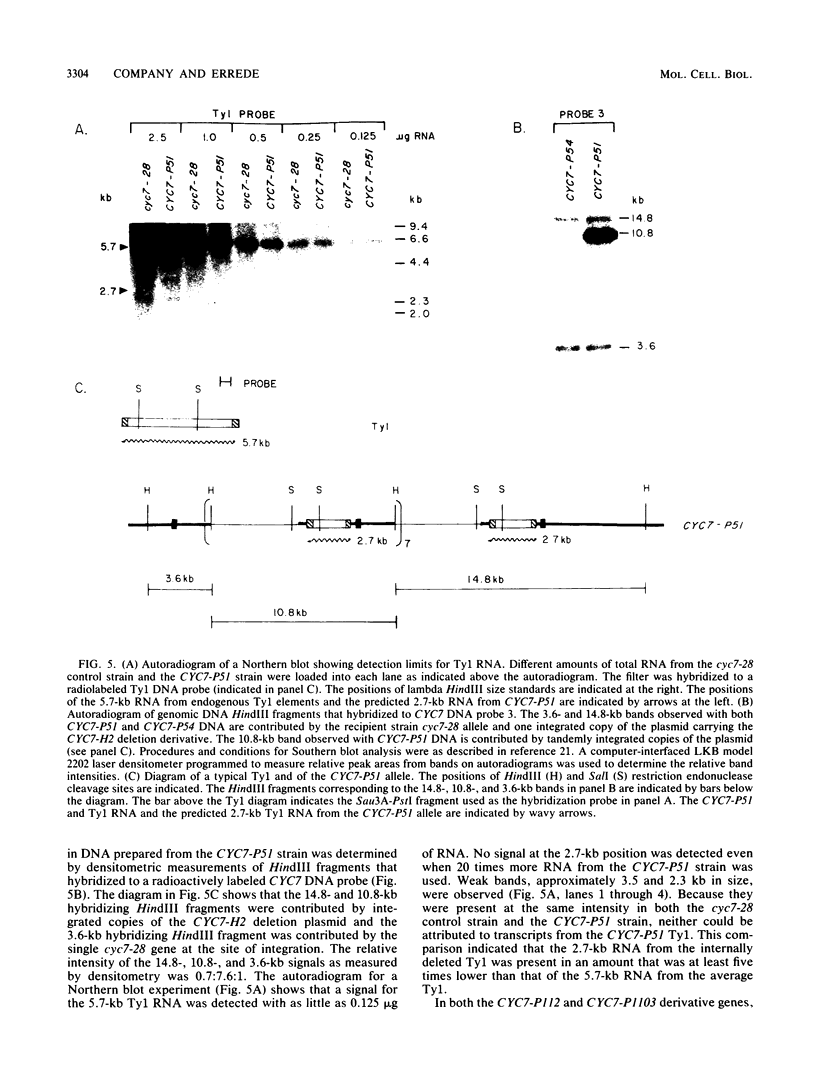
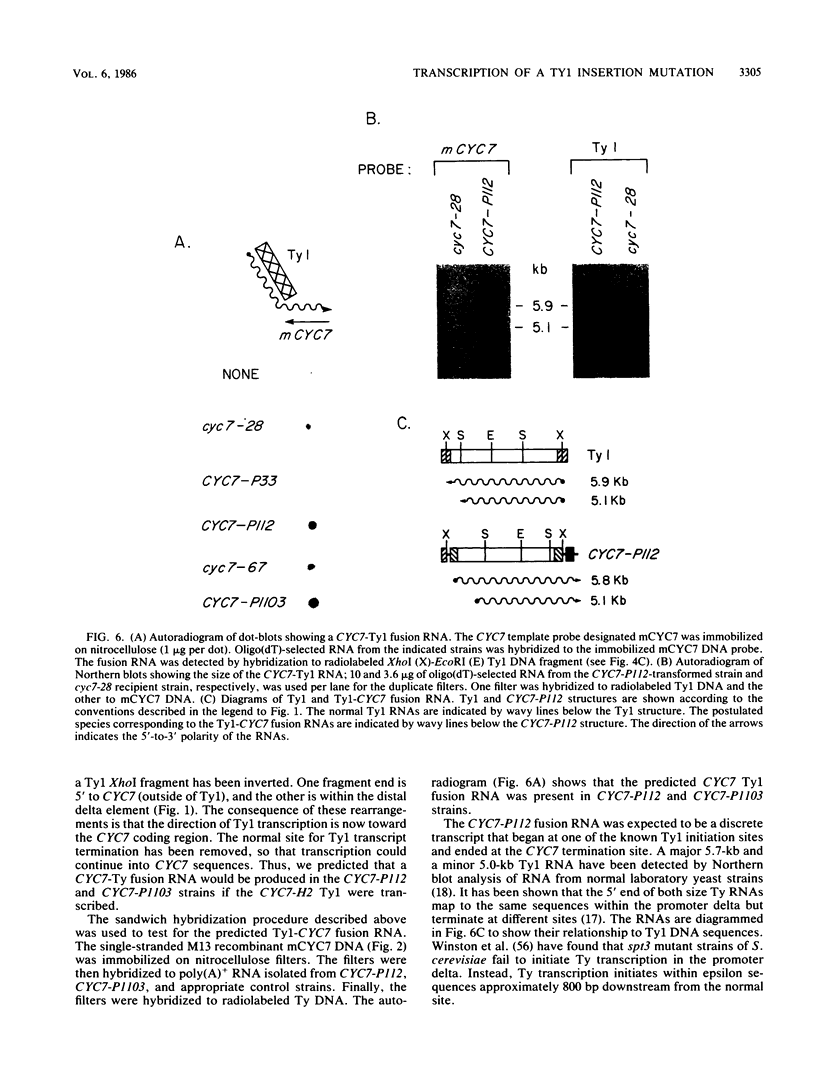
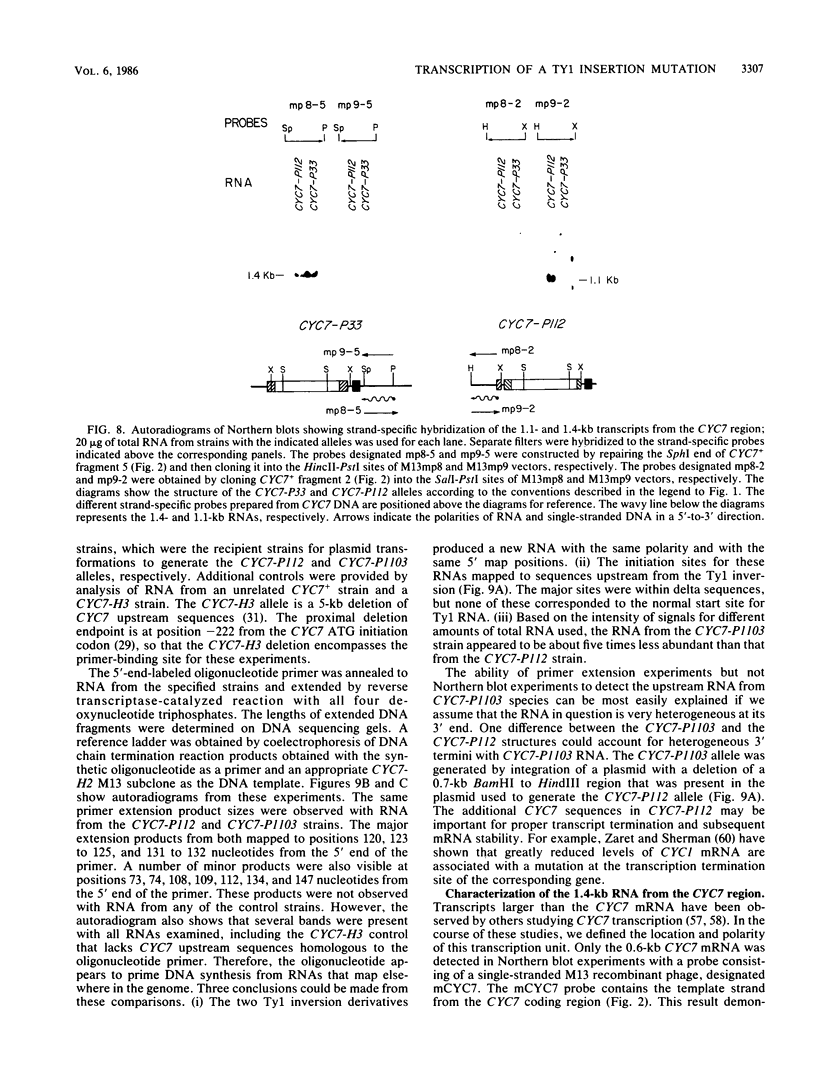
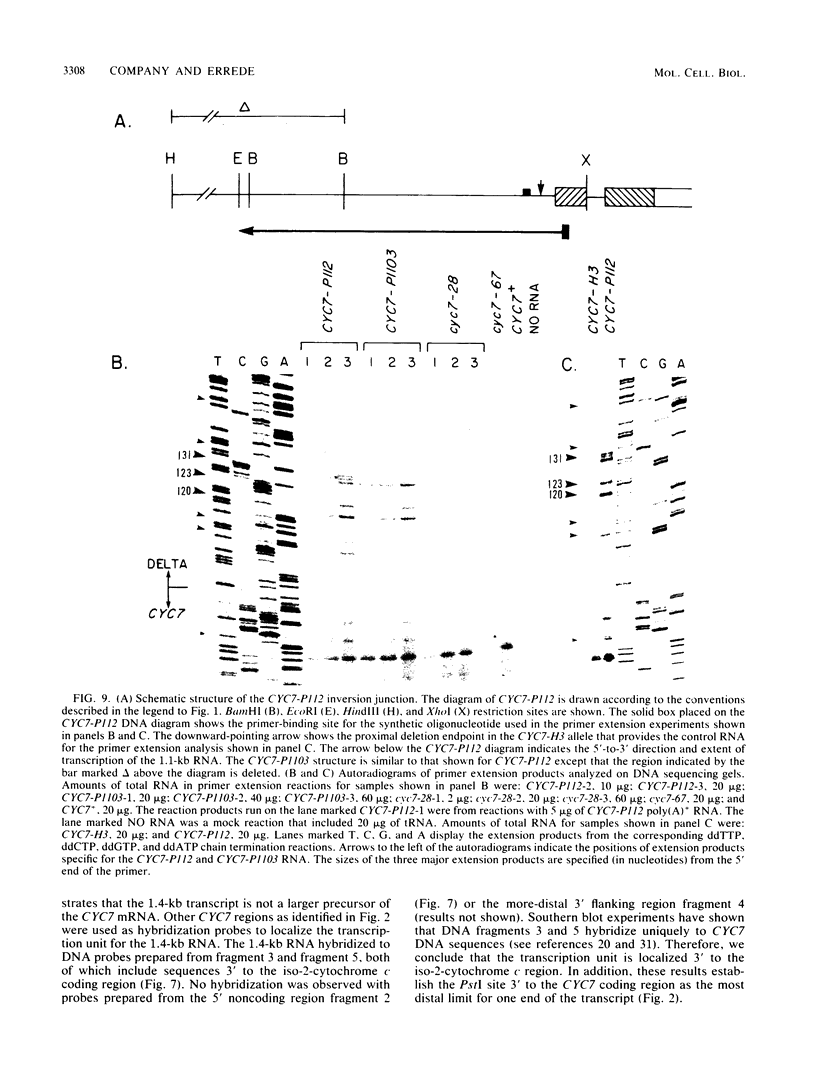
One class of Ty insertion mutation in Saccharomyces cerevisiae activates expression of adjacent structural genes. The CYC7-H2 mutation, in which a Ty1 element is inserted 5' to the iso-2-cytochrome c coding region of CYC7, causes a 20-fold increase in CYC7 expression. Deletion analysis of CYC7-H2 has shown that distal regions of the Ty1 element are not essential for the transcriptional activation at CYC7. In this report, we have analyzed Ty1 and CYC7 RNA from two CYC7-H2 deletion derivative genes to determine whether a direct correlation exists between transcription of Ty1 and transcription of the adjacent gene. Assuming that all Ty1 elements in the genome are transcribed equally, amounts of CYC7-H2 deletion derivative Ty1 RNA were found to be at least fivefold lower than the amount estimated for the average Ty1 element. These same Ty1 deletion derivatives caused a 20-fold increase in adjacent CYC7 expression. This finding suggests that the mechanism by which Ty1 activates adjacent gene expression does not require normal levels of Ty1 transcription. Two inversion derivatives of the CYC7-H2 Ty1 have also been analyzed. These derivatives did not produce any iso-2-cytochrome c or any normal CYC7 mRNA. Instead they were found to produce a Tyl-CYC7 fusion RNA. Consistent with our findings on CYC7-H2 Ty1 transcription, the amount of the fusion RNA was very low. In addition, the Ty1 inversion derivatives produced a new RNA that mapped to sequences upstream from the inverted Ty1 segment. Similar to Ty1 insertions that activate transcription, the new RNA was found to be transcribed away from Ty1.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach M. L. Ty1-promoted expression of aspartate transcarbamylase in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1984;194(3):395–401. doi: 10.1007/BF00425550. [DOI] [PubMed] [Google Scholar]
- Bingham P. M., Judd B. H. A copy of the copia transposable element is very tightly linked to the Wa allele at the white locus of D. melanogaster. Cell. 1981 Sep;25(3):705–711. doi: 10.1016/0092-8674(81)90177-x. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Garfinkel D. J., Styles C. A., Fink G. R. Ty elements transpose through an RNA intermediate. Cell. 1985 Mar;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Broach J. R., Atkins J. F., McGill C., Chow L. Identification and mapping of the transcriptional and translational products of the yeast plasmid, 2mu circle. Cell. 1979 Apr;16(4):827–839. doi: 10.1016/0092-8674(79)90098-9. [DOI] [PubMed] [Google Scholar]
- Cameron J. R., Loh E. Y., Davis R. W. Evidence for transposition of dispersed repetitive DNA families in yeast. Cell. 1979 Apr;16(4):739–751. doi: 10.1016/0092-8674(79)90090-4. [DOI] [PubMed] [Google Scholar]
- Carmichael G. G., McMaster G. K. The analysis of nucleic acids in gels using glyoxal and acridine orange. Methods Enzymol. 1980;65(1):380–391. doi: 10.1016/s0076-6879(80)65049-6. [DOI] [PubMed] [Google Scholar]
- Chaleff D. T., Fink G. R. Genetic events associated with an insertion mutation in yeast. Cell. 1980 Aug;21(1):227–237. doi: 10.1016/0092-8674(80)90130-0. [DOI] [PubMed] [Google Scholar]
- Clare J., Farabaugh P. Nucleotide sequence of a yeast Ty element: evidence for an unusual mechanism of gene expression. Proc Natl Acad Sci U S A. 1985 May;82(9):2829–2833. doi: 10.1073/pnas.82.9.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dretzen G., Bellard M., Sassone-Corsi P., Chambon P. A reliable method for the recovery of DNA fragments from agarose and acrylamide gels. Anal Biochem. 1981 Apr;112(2):295–298. doi: 10.1016/0003-2697(81)90296-7. [DOI] [PubMed] [Google Scholar]
- Dubois E., Jacobs E., Jauniaux J. C. Expression of the ROAM mutations in Saccharomyces cerevisiae: involvement of trans-acting regulatory elements and relation with the Ty1 transcription. EMBO J. 1982;1(9):1133–1139. doi: 10.1002/j.1460-2075.1982.tb01308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn A. R., Hassell J. A. A novel method to map transcripts: evidence for homology between an adenovirus mRNA and discrete multiple regions of the viral genome. Cell. 1977 Sep;12(1):23–36. doi: 10.1016/0092-8674(77)90182-9. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Control of eukaryotic messenger RNA synthesis by sequence-specific DNA-binding proteins. 1985 Aug 29-Sep 4Nature. 316(6031):774–778. doi: 10.1038/316774a0. [DOI] [PubMed] [Google Scholar]
- Eibel H., Philippsen P. Preferential integration of yeast transposable element Ty into a promoter region. 1984 Jan 26-Feb 1Nature. 307(5949):386–388. doi: 10.1038/307386a0. [DOI] [PubMed] [Google Scholar]
- Elder R. T., Loh E. Y., Davis R. W. RNA from the yeast transposable element Ty1 has both ends in the direct repeats, a structure similar to retrovirus RNA. Proc Natl Acad Sci U S A. 1983 May;80(9):2432–2436. doi: 10.1073/pnas.80.9.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder R. T., St John T. P., Stinchcomb D. T., Davis R. W., Scherer S., Davis R. W. Studies on the transposable element Ty1 of yeast. I. RNA homologous to Ty1. II. Recombination and expression of Ty1 and adjacent sequences. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):581–591. doi: 10.1101/sqb.1981.045.01.075. [DOI] [PubMed] [Google Scholar]
- Ernst J. F., Stewart J. W., Sherman F. The cyc1-11 mutation in yeast reverts by recombination with a nonallelic gene: composite genes determining the iso-cytochromes c. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6334–6338. doi: 10.1073/pnas.78.10.6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errede B., Cardillo T. S., Sherman F., Dubois E., Deschamps J., Wiame J. M. Mating signals control expression of mutations resulting from insertion of a transposable repetitive element adjacent to diverse yeast genes. Cell. 1980 Nov;22(2 Pt 2):427–436. doi: 10.1016/0092-8674(80)90353-0. [DOI] [PubMed] [Google Scholar]
- Errede B., Cardillo T. S., Teague M. A., Sherman F. Identification of regulatory regions within the Ty1 transposable element that regulate iso-2-cytochrome c production in the CYC7-H2 yeast mutant. Mol Cell Biol. 1984 Jul;4(7):1393–1401. doi: 10.1128/mcb.4.7.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errede B., Cardillo T. S., Wever G., Sherman F., Stiles J. I., Friedman L. R., Sherman F. Studies on transposable elements in yeast. I. ROAM mutations causing increased expression of yeast genes: their activation by signals directed toward conjugation functions and their formation by insertion of Ty1 repetitive elements. II. deletions, duplications, and transpositions of the COR segment that encompasses the structural gene of yeast iso-1-cytochrome c. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):593–607. [PubMed] [Google Scholar]
- Errede B., Company M., Ferchak J. D., Hutchison C. A., 3rd, Yarnell W. S. Activation regions in a yeast transposon have homology to mating type control sequences and to mammalian enhancers. Proc Natl Acad Sci U S A. 1985 Aug;82(16):5423–5427. doi: 10.1073/pnas.82.16.5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errede B., Company M., Swanstrom R. An anomalous Ty1 structure attributed to an error in reverse transcription. Mol Cell Biol. 1986 Apr;6(4):1334–1338. doi: 10.1128/mcb.6.4.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel D. J., Boeke J. D., Fink G. R. Ty element transposition: reverse transcriptase and virus-like particles. Cell. 1985 Sep;42(2):507–517. doi: 10.1016/0092-8674(85)90108-4. [DOI] [PubMed] [Google Scholar]
- Gruss P. Magic enhancers? DNA. 1984;3(1):1–5. doi: 10.1089/dna.1.1984.3.1. [DOI] [PubMed] [Google Scholar]
- Jauniaux J. C., Dubois E., Vissers S., Crabeel M., Wiame J. M. Molecular cloning, DNA structure, and RNA analysis of the arginase gene in Saccharomyces cerevisiae. A study of cis-dominant regulatory mutations. EMBO J. 1982;1(9):1125–1131. doi: 10.1002/j.1460-2075.1982.tb01307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laz T. M., Pietras D. F., Sherman F. Differential regulation of the duplicated isocytochrome c genes in yeast. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4475–4479. doi: 10.1073/pnas.81.14.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight G. L., Cardillo T. S., Sherman F. An extensive deletion causing overproduction of yeast iso-2-cytochrome c. Cell. 1981 Aug;25(2):409–419. doi: 10.1016/0092-8674(81)90059-3. [DOI] [PubMed] [Google Scholar]
- Mellor J., Fulton S. M., Dobson M. J., Wilson W., Kingsman S. M., Kingsman A. J. A retrovirus-like strategy for expression of a fusion protein encoded by yeast transposon Ty1. Nature. 1985 Jan 17;313(5999):243–246. doi: 10.1038/313243a0. [DOI] [PubMed] [Google Scholar]
- Mellor J., Malim M. H., Gull K., Tuite M. F., McCready S., Dibbayawan T., Kingsman S. M., Kingsman A. J. Reverse transcriptase activity and Ty RNA are associated with virus-like particles in yeast. Nature. 1985 Dec 12;318(6046):583–586. doi: 10.1038/318583a0. [DOI] [PubMed] [Google Scholar]
- Miller A. M., MacKay V. L., Nasmyth K. A. Identification and comparison of two sequence elements that confer cell-type specific transcription in yeast. Nature. 1985 Apr 18;314(6012):598–603. doi: 10.1038/314598a0. [DOI] [PubMed] [Google Scholar]
- Modolell J., Bender W., Meselson M. Drosophila melanogaster mutations suppressible by the suppressor of Hairy-wing are insertions of a 7.3-kilobase mobile element. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1678–1682. doi: 10.1073/pnas.80.6.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery D. L., Boss J. M., McAndrew S. J., Marr L., Walthall D. A., Zitomer R. S. The molecular characterization of three transcriptional mutations in the yeast iso-2-cytochrome c gene. J Biol Chem. 1982 Jul 10;257(13):7756–7761. [PubMed] [Google Scholar]
- Norgard M. V. Rapid and simple removal of contaminating RNA from plasmid DNA without the use of RNase. Anal Biochem. 1981 May 1;113(1):34–42. doi: 10.1016/0003-2697(81)90040-3. [DOI] [PubMed] [Google Scholar]
- Parkhurst S. M., Corces V. G. Forked, gypsys, and suppressors in Drosophila. Cell. 1985 Jun;41(2):429–437. doi: 10.1016/s0092-8674(85)80016-7. [DOI] [PubMed] [Google Scholar]
- Parnes J. R., Velan B., Felsenfeld A., Ramanathan L., Ferrini U., Appella E., Seidman J. G. Mouse beta 2-microglobulin cDNA clones: a screening procedure for cDNA clones corresponding to rare mRNAs. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2253–2257. doi: 10.1073/pnas.78.4.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne G. S., Bishop J. M., Varmus H. E. Multiple arrangements of viral DNA and an activated host oncogene in bursal lymphomas. Nature. 1982 Jan 21;295(5846):209–214. doi: 10.1038/295209a0. [DOI] [PubMed] [Google Scholar]
- Reeder R. H., Roan J. G., Dunaway M. Spacer regulation of Xenopus ribosomal gene transcription: competition in oocytes. Cell. 1983 Dec;35(2 Pt 1):449–456. doi: 10.1016/0092-8674(83)90178-2. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Roeder G. S., Fink G. R. DNA rearrangements associated with a transposable element in yeast. Cell. 1980 Aug;21(1):239–249. doi: 10.1016/0092-8674(80)90131-2. [DOI] [PubMed] [Google Scholar]
- Roeder G. S., Fink G. R. Movement of yeast transposable elements by gene conversion. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5621–5625. doi: 10.1073/pnas.79.18.5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder G. S., Rose A. B., Pearlman R. E. Transposable element sequences involved in the enhancement of yeast gene expression. Proc Natl Acad Sci U S A. 1985 Aug;82(16):5428–5432. doi: 10.1073/pnas.82.16.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M., Winston F. Identification of a Ty insertion within the coding sequence of the S. cerevisiae URA3 gene. Mol Gen Genet. 1984;193(3):557–560. doi: 10.1007/BF00382100. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J., Sherman F. Dependence on mating type for the overproduction of iso-2-cytochrome c in the yeast mutant CYC7-H2. Genetics. 1980 Apr;94(4):891–898. doi: 10.1093/genetics/94.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S., Mann C., Davis R. W. Reversion of a promoter deletion in yeast. Nature. 1982 Aug 26;298(5877):815–819. doi: 10.1038/298815a0. [DOI] [PubMed] [Google Scholar]
- Siliciano P. G., Tatchell K. Transcription and regulatory signals at the mating type locus in yeast. Cell. 1984 Jul;37(3):969–978. doi: 10.1016/0092-8674(84)90431-8. [DOI] [PubMed] [Google Scholar]
- Singh A., Sherman F. Deletions of the iso-1-cytochrome c and adjacent genes of yeast: discovery of the OSM1 gene controlling osmotic sensitivity. Genetics. 1978 Aug;89(4):653–665. doi: 10.1093/genetics/89.4.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. E. Form and function of retroviral proviruses. Science. 1982 May 21;216(4548):812–820. doi: 10.1126/science.6177038. [DOI] [PubMed] [Google Scholar]
- Williamson V. M., Cox D., Young E. T., Russell D. W., Smith M. Characterization of transposable element-associated mutations that alter yeast alcohol dehydrogenase II expression. Mol Cell Biol. 1983 Jan;3(1):20–31. doi: 10.1128/mcb.3.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson V. M., Young E. T., Ciriacy M. Transposable elements associated with constitutive expression of yeast alcohol dehydrogenase II. Cell. 1981 Feb;23(2):605–614. doi: 10.1016/0092-8674(81)90156-2. [DOI] [PubMed] [Google Scholar]
- Winston F., Durbin K. J., Fink G. R. The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell. 1984 Dec;39(3 Pt 2):675–682. doi: 10.1016/0092-8674(84)90474-4. [DOI] [PubMed] [Google Scholar]
- Wright C. F., Zitomer R. S. A positive regulatory site and a negative regulatory site control the expression of the Saccharomyces cerevisiae CYC7 gene. Mol Cell Biol. 1984 Oct;4(10):2023–2030. doi: 10.1128/mcb.4.10.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C. F., Zitomer R. S. Point mutations implicate repeated sequences as essential elements of the CYC7 negative upstream site in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Nov;5(11):2951–2958. doi: 10.1128/mcb.5.11.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]