Abstract
Meiotic recombination arises from Rec12/Spo11-dependent formation of DNA double-strand breaks (DSBs) and their subsequent repair. We identified Rec12-binding peaks across the S. pombe genome using chromatin immunoprecipitation after reversible formaldehyde cross-linking combined with whole-genome DNA microarrays. Strong Rec12-binding coincided with previously identified DSBs at the recombination hotspots ura4A, mbs1, and mbs2, and correlated with DSB formation at a new site. In addition, Rec12-binding corresponded to eight novel conversion hotspots and correlated with crossover density in segments of chromosome I. Notably, Rec12-binding inversely correlated with GC content, contrary to findings in S. cerevisiae. Although both replication origins and Rec12-binding sites preferred AT-rich gene-free regions, they seemed to exclude each other. We also uncovered a connection between binding sites of Rec12 and meiotic cohesin Rec8. Rec12-binding peaks lay often within 2.5 kb of a Rec8-binding peak. Rec12-binding showed preference for large intergenic regions and was found to bind preferentially near to genes expressed strongly in meiosis. Surprisingly, Rec12-binding was also detected in centromeric core regions, which raises the intriguing possibility that Rec12 plays additional roles in meiotic chromosome dynamics.
Introduction
Homologous recombination occurs in meiosis at high levels to ensure proper chromosome segregation during the first meiotic division, and to provide the basis for genetic diversity. Some regions in the genome show more recombination activity than others and are called hotspots. In Schizosaccharomyces pombe and Saccharomyces cerevisiae meiotic recombination at hotspots and elsewhere is initiated by DNA double-strand breaks (DSBs) in one of the two homologous chromosomes (Cervantes et al. 2000; Nicolas et al. 1989), usually at a number of positions across a range of 50 bp (Xu and Petes 1996) up to 2 kb (Cromie et al. 2005).
Chromatin structure can modulate the accessibility of a region for DSB formation, as shown for the histone acetylase mutant gcn5Δ at the meiotic recombination hotspot ade6-M26 in S. pombe (Yamada et al. 2004). Specific DNA sequences seem to have a minor role, as long as they are not required for DNA binding factors that in turn render the neighboring region accessible (α–hotspots). Despite of high overall sequence homology (98,6%) between the human and chimpanzee genomes, recombination hotspots, inferred by analyzing polymorphisms at orthologous loci and linkage disequilibrium, mapped at different locations in the two species (Winckler et al. 2005). In addition, the recombination frequency in human female autosomes is higher than in male autosomes (Kong et al. 2002; Mohrenweiser et al. 1998). Besides the well studied α–hotspots (Baudat and Nicolas 1997; Steiner et al. 2002), other hotspots exist, where alternative determinants render the chromatin amenable to DSB formation, e.g. nucleosome-excluding sequences (β–hotspots) (Kirkpatrick et al. 1999).
Meiotic DSBs are formed by the conserved topoisomerase II-related protein Spo11 (Rec12 in S. pombe), which works in a complex with several other proteins (Keeney 2001; Maleki et al. 2007). A Spo11 dimer forms phosphodiester links of its active site tyrosines, Y135, with the two 5′ ends of the DSB. The subsequent, staggered endonucleolytic incision (Neale et al. 2005), presumably catalyzed by the Mre11/Rad50/Xrs2 complex (MRX) and other activities, is inhibited in rad50S mutants in both yeasts (Alani et al. 1990; Cervantes et al. 2000; Young et al. 2002). This phenotype was employed in several, genome-wide analyses for Spo11-dependent DSBs in S. cerevisiae (Borde et al. 2004; Gerton et al. 2000; Mieczkowski et al. 2006). A prominent correlation of Spo11-dependent DSBs was found with high GC content. Correlation between recombination density and GC content was also found in the human genetic map (Yu et al. 2001), but not every hotspot displays high GC content (de Massy 2003). In accordance, an inverse correlation between preferred AT rich meiotic cohesin (Rec8) binding sites and DSB formation was reported in S. cerevisiae: Spo11 does not form DSBs in cohesin-binding regions (Glynn et al. 2004; Mieczkowski et al. 2006). Similarly, low Spo11 binding was found across budding yeast centromeres and telomeres, and in regions flanking the rDNA gene cluster. The absence of Spo11 matches the hypo-recombinogenic character of these specific regions. The transposition of the centromeric region CEN3 in S. cerevisiae leads to a decrease of gene conversion and crossing over at the novel centromere (Lambie and Roeder 1986; Lambie and Roeder 1988).
In this work, we have used the chromatin-immunoprecipitation method firstly to confirm Rec12 association with a meiotic recombination hotspot and secondly to globally map regions in the fission yeast genome where Rec12 binds. We could show that Rec12-binding agrees with recombination activity and DSB formation at a selection of Rec12-binding sites. Besides providing a map of Rec12-binding sites, we have identified a number of correlations between Rec12-binding sites and other chromosomal features.
Materials and methods
Strains, media and general genetic methods
Tetrad dissection was described by (Baur et al. 2005). Standard minimal medium (MMA), yeast extract media (YEL) and (YEA), malt extract media (MEA), and the general genetic methods were described by (Moreno et al. 1991). Synthetic medium (EMM) with 2% glucose and (EMM-N) with 1% glucose were described in (Nurse 1975). Strains used are listed in Table 1. Rec12 was tagged at the C-terminus with 13 myc epitopes at its endogenous chromosomal locus by PCR-based gene targeting (Bähler et al. 1998). The resulting strain was wild type with respect to spore viability and intragenic recombination at ura4A. The ORFs of rec6 and rec14 were replaced by the hygromycin resistance cassette to create the deletion mutants rec6Δ::hygR and rec14Δ::hygR, respectively (Hentges et al. 2005). The deletion strains showed spore viabilities less than 32 % and intragenic recombination at ura4A was reduced more than 2000-fold compared to wild type. Tagging and gene disruption constructs were confirmed by PCR and subsequent sequencing. The tagged Rec12 fusion protein was detected by Western analysis in meiotic extracts of a pat1-114 strain.
Table 1.
S. pombe strains
| Strain | Genotype | Derivation |
|---|---|---|
| KLY52 | h+ ura4A ura4-D18 rec12myc::kanR pat1-114 | this study |
| KLY59 | h− ura4-D18 rec12myc::kanR | this study |
| KLY118 |
h+ ura4A ura4-D18 rad50S pat1-114
rec12myc::kanR |
this study |
| KLY108 |
h+ ura4A-10 ura4-D18 rad50S pat1-114
rec14Δ::hygR rec12myc::kanR |
this study |
| KLY123 |
h+ ura4A ura4-D18 rad50S pat1-114 rec6Δ::hygR
rec12myc::kanR |
this study |
| KLY288 | h+ gpd1-E13 | H. Aiba |
| GP3718 | h+ ade6-M3049 end1-458 pat1-114 rad50S | G. Smith |
| 145-5768 | h− sod2::ura4 ura4-D18 | Bern |
| 144-5755 | h+ lys3-37 ura4-D18 | Bern |
| 3-97 | h− lys3-97 | Bern |
| 4-127 | h+ ura1-61 | Bern |
| 3-120 | h− pro1-1 | Bern |
| 1-15 | h+ ade3-58 | Bern |
| 3-93 | h− lys1-131 | Bern |
| 2-69 | h+ his1-102 | Bern |
| 152-6045 | h− cdc1-7 | Bern |
| 29-1147 | h+ cdc1-7 | Bern |
| 3-87 | h− leu2-120 | Bern |
| 4-150 | h+ lys7-2 | Bern |
| 51-2020 | h+ cdc3-6 | Bern |
| 51-2021 | h− cdc3-6 | Bern |
| 29-1149 | h− cdc13-117 | Bern |
| 3-99 | h− lys4-95 | Bern |
| 3-100 | h+ lys4-95 | Bern |
| 15-588 | h− lys4-8 | Bern |
| 15-589 | h+ lys4-8 | Bern |
| 27-1045 | h+ ade10-13 | Bern |
| 27-1046 | h− ade10-13 | Bern |
| 154-6151 | h− clr6-1 | Bern |
| 156-6226 | h+ sme2Δ::ura4+ ura4-D18 | Bern |
| 62-2444 | h− ade6-469 ura4-D18 | Bern |
Beside the auxotrophies lys4-8 (−95) and ade10-13, the following genetic markers (with respective phenotypes) were used for tetrad analysis: gpd1-E13 (no growth on YEA + 1.6 M sorbitol (Ohmiya et al. 1995)), rec12myc::kanR (growth on YEA + 100 mg/ml G418), cdc13-117, clr6-1, and cdc3-6 (no growth on YEA at 37°C). All crosses involved two unlinked, heterozygous markers, except for sme2Δ::ura4, where sme2Δ::ura4 ura4-D18 was crossed with ura4-D18.
Chromatin Immunoprecipitation
Haploid pat1-114 strains were grown in YEL at 25°C to saturation (OD595 >5). The preculture was diluted into EMM and grown at 25°C to OD595 of 0.4. Cells were harvested by centrifugation, washed once with one volume water, resuspended in one volume of EMM-N (EMM without nitrogen source), and kept for at least 16 hours at 25°C to arrest cells in G1. NH4Cl was added to 0.5 g/l, and the temperature risen to 34°C to start meiosis. At various time points, 1 ml samples were taken for DAPI staining, and 50 ml samples for chromatin immunoprecipitation. Formaldehyde solution (33.3%, Fluka 47629) was added to a final concentration of 1%, followed by incubation for 15 minutes at room temperature on a wheel. Due to massif meiotic protein degradation, we were not able to recover precipitated DNA from un-fixed cells even in the presence of multiple, highly concentrated protein inhibitors. Cells were washed with one volume of cold PBS (130 mM NaCl, 10 mM NaPO4, pH 7.5), and cell pellets frozen in liquid N2.
Cells were lysed in 400 μl of lysing buffer (50 mM HEPES pH 7.5, 140 mM NaCl, 1mM EDTA, 1% (w/v) Triton X-100, 0.1% (w/v) Na desoxycholate, 1mM Pefabloc SC, 0.5% (v/v) Pefabloc SC Protector Solution, 0.5% (v/v) n-octanol, and protease inhibitor cocktail from Roche (1 big tablet/25 ml buffer)) with acid-washed glass beads using the FastPrepMachine (BIO101) at level 6.5 for 3 cycles, 40 seconds each. Tubes were kept on ice between cycles. Crude extract was collected, and the beads were washed with 300 μl of lysing buffer. Pooled extract and wash was sonicated with a micro tip (tapered 1/8″) in a Branson digital Sonifier 250 for 40 cycles at 35% amplitude (pulse on: 0.9 seconds, pulse off: 7 seconds) to fragment the chromatin to an average size of 650 bp.
To obtain whole cell extracts (WCE) the samples were centrifuged once for 5 minutes, and the supernatant (SN) again for 15 minutes at 13′000 rpm in a cold micro centrifuge. 500 μl of WCE was precipitated for 3 hours at 4°C on a rotation wheel with 30 μl of pre-coated magnetic beads per sample (3 × 107 Dynal M-280 beads, Sheep anti-mouse IgG). The beads had been coated with 15 μl monoclonal mouse anti-myc antibodies (c-myc (9E10):sc-40, from Santa Cruz) over night at 4°C in PBS buffer containing 5 mg/ml BSA. Using the MPC-E magnet (Dynal), the beads were collected, drained from solution, and washed with 1 ml of the following buffers: twice with lysing buffer, twice with washing buffer A (50 mM HEPES pH 7.5, 500 mM NaCl, 1 mM EDTA, 1% (w/v) Triton X-100, 0.1% (w/v) Na desoxycholate, and protease inhibitors), twice with washing buffer B (10 mM Tris HCl pH 8.0, 250 mM LiCl, 1 mM EDTA, 0.5% (w/v) NP-40, 0.5% (w/v) Na desoxycholate, and protease inhibitors), and once with TE (10 mM Tris HCl pH 8.0, 1 mM EDTA). 50 μl elution buffer (50 mM Tris HCl pH 8.0, 10 mM EDTA, 1% (w/v) SDS) was then added to the drained beads, and the precipitates were released at 65°C during a 10 minutes incubation with occasional vortexing. After a brief centrifugation at 13′000 rpm at room temperature, the SN was transferred to a fresh tube and 150 μl TE/SDS (10 mM Tris pH 8.0, 1 mM EDTA, 1% (w/v) SDS) was added (ChIP). Also, 5 μl of retained WCE was added to 195 μl TE/SDS (input). After incubating both ChIP and input at 65°C over night for reversal of cross-linking, proteins were digested with 1 mg/ml Proteinase K (Invitrogen) in TE at 50°C for 2 hours. After adding glycogen (final concentration of 0.4 mg/ml), the DNA was purified by two phenol/chloroform/isoamylalcohol-extractions and one chloroform/isoamylalcohol-extraction in PGL light tubes (Eppendorf). The DNA was finely resuspended in 30 μl TE containing 0.05 mg/ml Rnase A (Dnase-free, QbioGene) and incubated at 37°C for 30 minutes. DNA concentration was measured with the NanoDrop device.
Real-time PCR
Real-time PCR was carried out with AmpliTaq Gold® DNA Polymerase (TaqMan® Universal PCR Master Mix, Applied Biosystems). TaqMan probes used were RTKL1 (forward primer at a final concentration of 18 μM: 5′-CAAAGTTATGGATGCTAGAGTATTTCAAAGC-3′, reverse primer at a final concentration of 18 μM: 5′- TCGACCGCGACTGACAAG-3′, labeled primer at final concentration of 5 μM: 5′FAM-CTTGGCAATGGGATTTT-NFQ3′). The labeled primer localizes approximately 450 bp distal to the mapped DSB site of ura4A (Gregan et al. 2005). PCR steps for the ABI prism 7000 were 2 min at 50°C, 10 min at 95°C, followed by 40 cycles of 15 sec at 95°C, and 1 min at 60°C. Duplets of 1-5 μl (50 ng) of immunoprecipitated or input DNA were analyzed per sample. The amount of template in one μl of input DNA was set to 100%. The precipitated templates in the ChIP samples were given in percent of input.
DNA labeling, microarray hybridization, and data processing
The labeling, hybridization procedures, and data processing were described elsewhere (Heichinger et al. 2006). Two hybridizations were carried out simultaneously on both microarrays (ORF and intergenic), another two hybridizations were probed only on the intergenic microarray. In three hybridization experiments, we labeled 300 to 600 ng immunoprecipitated DNA with Cy5 and similar amounts of input DNA with Cy3. In the forth experiment, we swapped the dye. The mean data of these 4 hybridization experiments were used for further calculations. Four independent control experiments with untagged Rec12 repeatedly gave no labeling of the mock-immunoprecipitated DNA sample, whereas the input DNA was labeled fine. Information on probes indicated in the text is available from a searchable Microarray Primer Database (Penkett et al. 2006). All raw data will be available from ArrayExpress. In addition, all processed data will be available from: http://www.sanger.ac.uk/PostGenomics/S_pombe/. To identify Rec12 binding peaks, the sliding-window approach of the ChIPOTIe software (version 1.0) was used on the log2 values of the average of two (ORF probes) or four (intergenic probes) Rec12-binding ratios with a sliding window of 1000 bp and 250 bp steps. Prism software version 4.0 was used for calculations of correlation tests.
The transformation of the Affimetrix data set (meiotic cohesin binding, see (Ding et al. 2006)) to our intergenic and ORF chip architecture was done by location of the Affimetrix oligos by sequence (http://chromosomedynamics.bio.titech.ac.jp/download/pombe_chip/pombe_chip.html), and assigning the Rec8 to input ratio from the 250 bp window given in (Ding et al. 2006) for the wild type to every oligo. These values, assigned to the sequence, were then used for calculation of the average over the length of our intergenic and ORF probes.
DSB Southern
5 × 108 cells from a meiotic pat1-114 rad50S time course were harvested at appropriate time points (T0, T2, T3, T4, T5, T6), washed with 50 mM EDTA pH 8.0, and resuspended in 300 μl SpheroBuffer (0.4 M EDTA, 10 mM NaN3, 5 mM DTT, 1 mg/ml zymolyase (Seikagaku 120493), 5 mg/ml lysing enzyme (Sigma L-1412), and embedded in low melting-point agarose to a final concentration of 1%. Cells in agarose plugs were lysed in 1.8 ml SpheroBuffer at 37°C for one hour, proteins were digested once in 1.8 ml NDS/PK (0.5 M EDTA, 10 mM Tris pH 7.5, 10 mM NaN3, 1 % Na lauryl sarcosine, 1 mg/ml Proteinase K) at 50°C over night, and again in 1.8 ml SDS/PK (0.5 M EDTA, 10 mM Tris pH 7.5, 0.5 % SDS, 1 mg/ml Proteinase K) at 50°C over night. Plugs were extensively washed with TE containing 1mM PMSF, exchanging the buffer several times, at room temperature and stored afterwards in TE at 4°C.
Genomic DNA embedded in agarose plugs was digested in the appropriate buffers with restriction enzymes according to the supplier (NEB). In particular, in 200 μl final volume 100 units ClaI were used at 37°C over night. Melted agarose plugs were loaded on a 0.8% TBE (90 mM Tris base, 90 mM boric acid (pH 8.3), 2mM Na2EDTA) agarose gel, and run over night in the cold room. After ethidium bromide staining, the DNA was blotted on GeneScreenPlus membrane (PerkinElmer) with the VacuGeneXL device (Pharmacia), according to the manufacturer. Radioactive probes were made by random priming of gel-purified PCR fragments with [a-32P]dCTP using the Ready-To-Go DNA Labeling Beads (Amersham). Hybridization and membrane washing procedures were done according to the membrane supplier. Parental and DSB bands were detected by a phosphoimager, quantified with the AIDA software (Raytest, Germany), and related to each other after background subtraction by setting the parental band to 100%.
Results
Rec12 associates with the meiotic DNA break region at the ura4A hotspot
Point mutations in the 5′ region of the ura4A gene displayed gene conversion frequencies of 18%, whereas in the 3′ region only 6% was detected (Baur et al. 2005). This conversion gradient was explained by initiation of recombination in the 5′ upstream region of ura4A by DSB formation. A meiotic DSB site has been detected and mapped approximately 500 bp upstream of the ura4A translation start codon (Gregan et al. 2005; Sakem 2005).
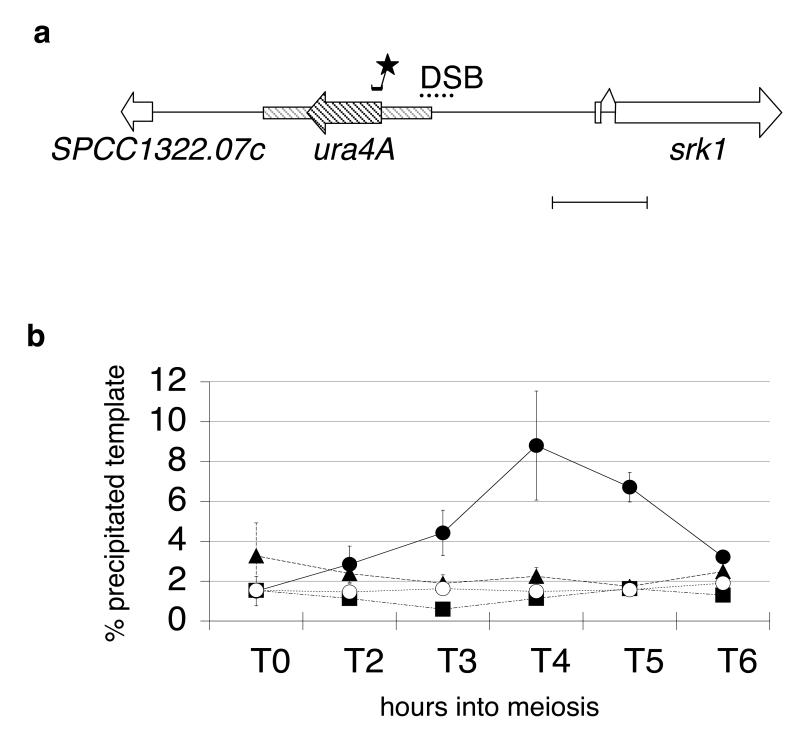
To find out, whether Rec12 associates with sequences in the DSB formation region of ura4A, we employed chromatin immunoprecipitation (ChIP) with a functional, C-terminal myc-tagged Rec12 in a rad50S mutant. The experiments were carried out by induction of meiosis in haploid pat1-114 mutant strains with the advantage of high synchronicity (Beach et al. 1985). pat1-114 meiosis shows a chromosome segregation defect in the first meiotic division (Yamamoto and Hiraoka 2003), but has been used systematically for the timing of early meiotic events including DSB formation (Cervantes et al. 2000; Young et al. 2002). In addition, our strains carried the rad50S mutation in which DSBs remain un-resected (Young et al. 2002). Rec12myc-DNA complexes fixed by formaldehyde were purified from meiotic cells by immunoprecipitation using an anti-myc antibody. Precipitated DNA was tested for ura4A sequences by real time PCR (Fig. 1). Coincident with DSB formation, the association of Rec12 with ura4A was found to increase from the time-point 0 hours (1.6%) to 4 hours (9%).
Fig. 1.
Rec12 binds to ura4A hotspot sequences
a. Map of the ura4A insertion used for real time PCR detection of Rec12-binding sequences. ORFs are given as horizontal arrow boxes. The 1.8 kb HinD III ura4 fragment (Grimm et al. 1988), inserted between SPCC1322.07c and srk1, is cross-hatched. Dots indicate the region of DSB formation (Gregan et al. 2005). The location of the ura4A TaqMan® probe is given by a small bar with an asterisk. The length of the scale bar is 1 kb.
b. Graph showing the ratio of real time PCR product from immunoprecipitated chromatin versus total chromatin in rec12myc rad50S (filled circle), rec14Δ rec12myc rad50S (filled triangle), rec6Δ rec12myc rad50S (filled square), and rad50S rec12+(open circle) strains, during meiotic progression in a haploid pat1-114 strain. Independent experiments were performed four times for wild type, three times for rec14Δ, and twice for rec6Δ. Experiments with the untagged control strain were repeated twice. The y-axis gives the ratio of real time template of immunoprecipitated sample to input sample, normalized to 1 μl input, which was set to 100% (see Material and methods). Error bars indicate the standard error of the mean (SEM).
Strains with rec14Δ or rec6Δ deletions showed a strong decrease in meiotic recombination (Evans et al. 1997) and a rec14 mutant had the same altered linear elements as a rec12 mutant (Molnar et al. 2003). No more ura4A hotspot sequences were precipitated in rec14Δ and rec6Δ strains than in a rec+ strain with untagged Rec12 (Fig. 1B), indicating a requirement of Rec14 and Rec6 for formation of DSBs.
Global analysis of Rec12-binding
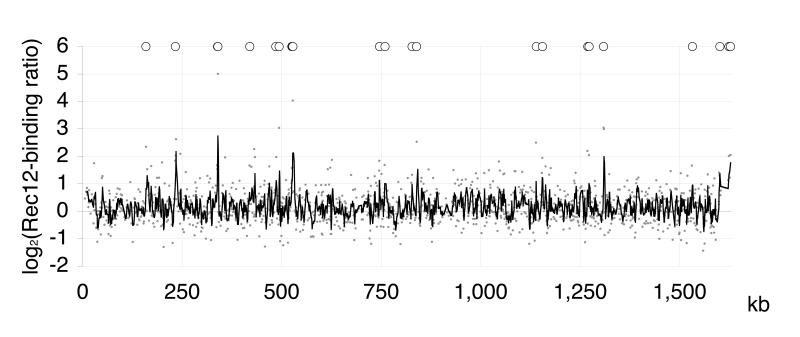
The capability of Rec12myc to associate with ura4A hotspot sequences prompted us to precipitate Rec12myc-DNA sequences from total chromatin to construct a genomic Rec12-binding map by hybridization to DNA microarrays. The microarrays covered all coding and non-coding regions of the S. pombe genome on probes of 180-500 bp (ORF array) (Lyne et al. 2003) and 36 −2900 bp (intergenic array) (Heichinger et al. 2006). Rec12myc immunoprecipitated DNA as well as input DNA from time-point 4 hours were differentially labeled and hybridized simultaneously to the microarrays. The Rec12-binding ratio was defined as the coefficient of the amount of immunoprecipitated DNA to input DNA on each probe. Data for the left arm of chromosome II from four independent hybridization experiments are shown in Fig. 2.
Fig. 2.
Mapping of Rec12-binding on the left arm of chromosome II
On the x-axis, the left telomere is given at 0 kb, the centromere at 1.600 kb. Microarray signal ratios of Rec12 enrichment relative to input were plotted for each probe against chromosome position and are given in a log2 scale as the mean of 2 (ORF sequences) or 4 (intergenic sequences) independent experiments (small gray dots). The black line indicates a moving average curve over 3 values (~3.3 kb). The Rec12-binding peaks determined by the CHIPOTIe algorithm are indicated above the curve (O).
To single out significant Rec12-binding peaks, the ChIPOTIe sliding average algorithm for uniformly arranged data was applied (Buck et al. 2005). In this way, 144 Rec12-binding peaks were identified genome-wide (Table S1; Fig. 2).
DSB formation near a Rec12-binding peak
To check for DSB formation near a strong Rec12-binding peak, SPBC1685.13 on chromosome II (Rec12-binding ratio on probe chr2.187.1 >16, Table S1), Cla I digested genomic DNA from time-course samples of haploid pat1-114 rad50S meiosis was analyzed using Southern hybridization (Fig. 3). Three meiosis-specific bands could be detected at 4 hours into meiosis and the sum was calculated to be 8.1% compared to the parental Cla I band. Thus, at sites where meiotic DSBs are formed, high level of Rec12-binding is found on a nearby probe.
Fig. 3.
Meiotic DSBs appear at a Rec12-binding peak
A Southern blot of Cla I digested genomic DNA from a pat1-114 meiotic time-course is shown. Above the autoradiogram, hours into meiosis are indicated. The arrows on the right side indicate meiosis-specific DSB fragments, and their quantification with respect to the parental Cla I fragment. The non-meiosis specific band below may arise from a cross-hybridization. Such bands were also observed at other loci (Cromie et al. 2006). The sketch on the right gives the chromosomal location of the three meiosis-specific DSBs on chromosome II (bolts). The arrowed (ORF) and gray (intergenic) boxes give the corresponding probes on the microarrays, together with the probe names. The bar with an asterisk indicates the location of the radioactive probe.
Hotspots of recombination localize to sites with high Rec12-binding
To correlate Rec12-binding with recombination activity, probes with various Rec12-binding ratios were checked for nearby genetic markers. Among eight markers with considerably high Rec12-binding ratios on neighboring probes were two CHIPOTIe Rec12-binding peaks, chr2.1694.1 and chr2.103.1 (Table S1)
From crosses of various combinations of heteroallelic markers, tetrads were dissected (Table 2) and the spore clones analyzed for the respective phenotypes (Material and Methods). Gene conversion frequencies ranging from 1 % to 16.7 % clearly exceeded the average NMS frequency of 0.28% for all studied genes (Gygax 1984; Munz 1994), and for six markers the values were even higher than the value for the well studied meiotic recombination hotspot ade6-M26 (5.1%) (Gutz 1971).
Table 2.
Increased gene conversion frequencies near Rec12-binding sites
| Rec12-binding ratio a (probe) |
Marker | Chromosome | Conversions | Tetrads analyzed |
Conversion frequency (%) |
||
|---|---|---|---|---|---|---|---|
| 3+:1− | 1+:3− | 4+:0− | |||||
| 32.1 (chr2.103.1) | sme2Δ::ura4 | II | 0 | 1 | 0 | 100 | 1.0 |
| 30.1 (chr2.1694.1) | gpd1-E13 | II | 11 | 0 | 1b | 108 | 12.0 |
| 3.6 (chr1.759.1) | rec12myc::kanR | I | 13 | 5 | 0 | 108 | 16.7 |
| 3.1 (chr2.139.1) | cdc13-117 | II | 11 | 7 | 0 | 220 | 8.2 |
| 2.5 (SPBC1105.02c) | lys4-8 | II | 2 | 11 | 0 | 207 | 6.3 |
| 2.5 (SPBC1105.02c) | lys4-95 | II | 1 | 6 | 0 | 56 | 12.5 |
| 2.3 (chr2.310.1) | clr6-1 | II | 3 | 0 | 0 | 37 | 8.1 |
| 2.1 (chr1.1107.1) | cdc3-6 | I | 8 | 10 | 0 | 141 | 12.8 |
| 1.4 (chr3.475.1) | ade10-13 | III | 0 | 2 | 0 | 135 | 1.5 |
Average Rec12-binding ratio of four (intergenic probes) or two (ORF probes) experiments.
One tetrad with two conversion events was found, and counted twice.
Rec12-binding and crossover on chromosome I
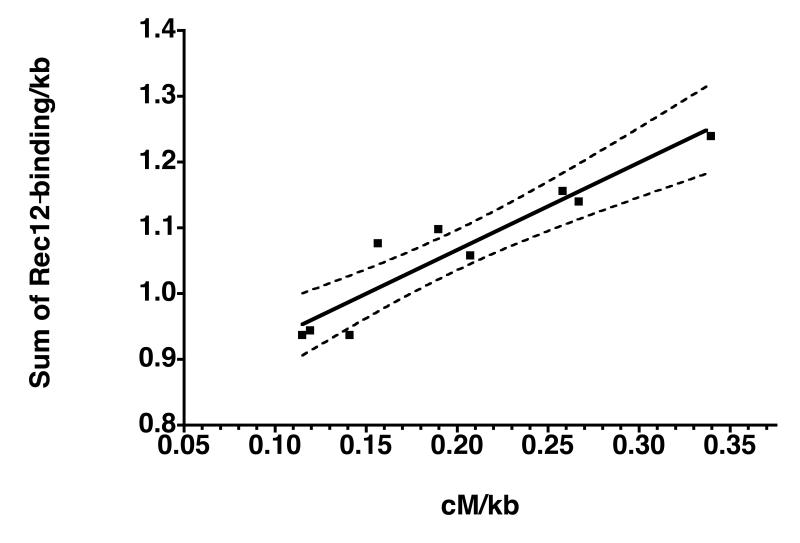
Genetic distance between two linked markers is a function of the number of crossovers in the interval. As crossover is one way to resolve recombination intermediates initiated at Rec12-dependent DSBs, a positive correlation between Rec12-binding and genetic distance would be expected. We used a set of different markers, which map across chromosome I (Fig. S2), and measured intergenic recombination in the intervals (Table S2). The genetic distance (cM/kb) was compared with the sum of Rec12-binding ratios, which was adjusted to the sum of the probe lengths in the interval. The two sets of data were strongly correlated (Pearson coefficient: 0.94, p-value =0.0002). Fig. 4 shows the correlation plot with a simple linear regression fit. The sum of background binding due to unspecific DNA precipitation with the antibody over the long intervals could be responsible for such a positive correlation, although no background binding was detected in control experiments with untagged Rec12 (Material and Methods). To test whether the correlation depends on background binding, the Rec12-binding ratios were randomly redistributed over chromosome I and added up again in the same intervals (Table S3). Less than 5% of the randomizations showed Pearson correlation p-values lower than the observed p-value. If the correlation would be a consequence of background binding, more randomizations with p-values similar to the observed p-value would be expected. We conclude that the amount of Rec12-binding per unit DNA length is not evenly distributed along chromosome I, and that high Rec12-binding is associated with increased recombination.
Fig. 4.
Rec12-binding correlates with crossovers
Correlation plot of genetic distance in cM/kb measured in intervals along chromosome I (see Table S2) versus the sum of the Rec12-binding ratios in the same intervals divided by the sum of probe lengths. The linear correlation line (solid line) is given together with its 99 % confidence limit (dashed lines).
Distribution of Rec12-binding peaks with respect to chromosomes, large intergenic regions, non-protein coding genes, and centromeres
From the 144 CHIPOTIe Rec12-binding peaks (Table S1), 69 were on chromosome I, 44 on chromosome II, and 31 on chromosome III. If the hotspots would be evenly distributed, one would expect less hotspots on chromosomes I and III (64 and 28) but more on chromosome II (52). However, the observed distribution is not significantly different from the expected one (χ2 = 1.01, p = 0.6). Surprisingly, the average Rec12-binding ratio on chromosome II (1.24) is higher than on chromosome I (1.18) and III (1.16, see Table S4). Nevertheless, the ratio of genetic length versus physical length is the same on chromosomes I and II (0.17 cM/kb) and higher on chromosome III (0.22 cM/kb) (Munz 1994). The higher average Rec12-binding ratio and the lower numbers of Rec12-binding peaks may reflect more evenly distributed Rec12 on chromosome II than on the other chromosomes. Extended “cold” or “hot” regions were not detected, in difference to S. cerevisiae (Gerton et al. 2000).
The major meiotic DSB sites mbs1 and mbs2 localize to so called “gene-free regions” (Cromie et al. 2005). These 4 to 8 kb long sequences occur on all three chromosomes (Wood et al. 2002). They display disproportional spreading of the four nucleotides, sometimes accompanied by repeats (low complexity), and are often associated with (G − C)/(G + C) strand switches (Lobry 1996), where a region with prominent (G) content is followed by a region of similar size with prominent (C) content. Besides the 51 low-complexity gene-free regions, another 21 gene-free regions of similar size lacking low complexity exist. 17 out of the 144 CHIPOTIe Rec12-binding peaks fall into gene-free regions (Table S1), among them mbs2 (measured on probe chr1.279.2) with a Rec12-binding ratio of 4. Comparison of the total length of all large gene-free region probes (280.5 kb) with the total length of all measured probes (12′064 kb) leads to an expectation of only 3 hotspots. Thus, Rec12 clearly shows a preference to bind in large gene-free regions. The Rec12-binding ratios on probes within gene-free regions had a higher average value (1.54) compared to the average value of all probes in intergenic regions (1.21, Fig. 5). The effect was most prominent on chromosome I, although two large gene-free regions had low Rec12-binding ratios (data not shown). The same trend was detected on chromosomes II and III, but with overlapping standard errors (Fig. 5).
Fig. 5.
Rec12 binds to large gene-free regions particularly on chromosome I
The average Rec12-binding ratio for the probes in large gene-free regions (dark bars) versus the average for all probes in intergenic regions (light bars) is shown. The ratios are given for the three chromosomes separately, and globally (on the right). The bars indicate the standard error of the mean. Large gene-free regions of the three chromosomes do not vary in length (classes of increasing 1′000 bp steps from 2′000 bp to 9′000 bp, χ2 = 20.6, p=0.1 in 95% confidence interval).
13 Rec12-binding peaks were found near non-protein coding genes (see Table S1). These genes are transcribed into poly (A)-bearing RNAs but lack long open reading frames. Based on the total length of all annotated non-protein coding genes (185 kb), only two Rec12-binding peaks were expected. The average of Rec12-binding to probes covering annotated non-protein coding genes (1.59 ± 0.24) was higher than the average of all probes (1.20 ± 0.01). It has been pointed out, that non-protein coding genes often map to intergenic regions longer than 2.4 kb and 20 % of them are in large gene-free regions (Watanabe et al. 2002). Out of the 17 large gene-free regions with a Rec12-binding peak, only 4 contain an annotated non-protein coding gene (24 %). Thus, the Rec12-binding correlations with large gene-free regions and non-protein coding genes may be independent. Alternatively, there may be so far undetected non-protein coding genes in large gene-free regions. It has been extrapolated that about 300 such genes remain to be discovered (Watanabe et al. 2002).
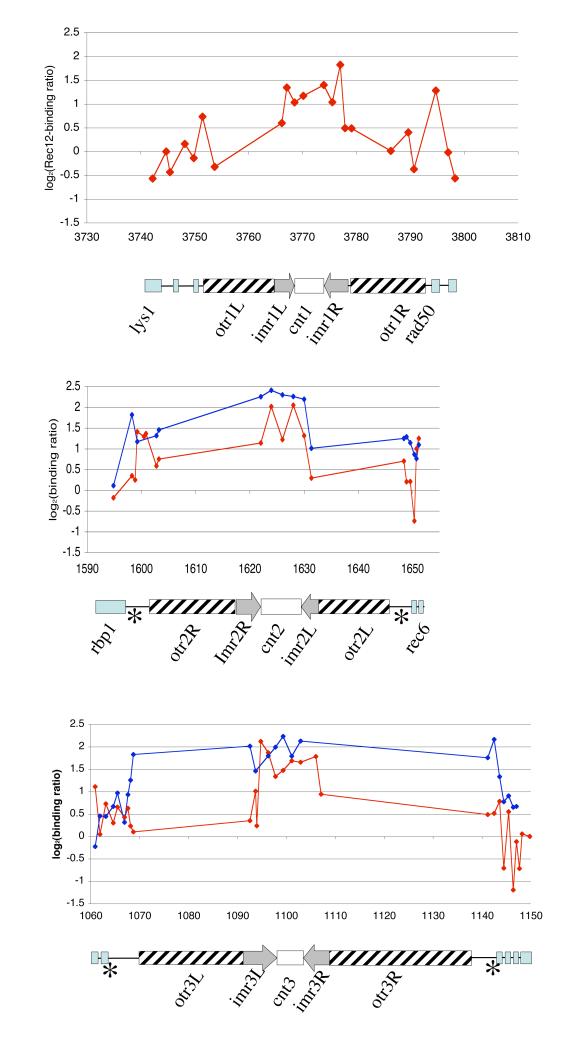
Recombination across centromere II was reported to be low (Nakaseko et al. 1986). Unexpectedly, strong Rec12-binding was detected at the inner core and inner repeats of all three centromeres (Fig. 6). The ChIPOTIe algorithm captured these sites as Rec12-binding peaks (Table S1).
Fig. 6.
Rec12-and Rec8-binding in centromere regions
The averaged Rec12-binding ratios (red dots) from four experiments (intergenic probes) or two experiments (ORF probes) and the transformed Rec8-binding ratios (blue dots) from the Hiraoka laboratory (Ding et al. 2006) are plotted against chromosomal location. The units on the x-axis are in kb and scaled from the left chromosome ends, the centromere structures are depicted below. Light blue boxes indicate ORF probes, cross-hatched boxes the outer repeats, light gray arrows the inverted inner repeats, asterisks tRNA clusters, and white boxes the core regions. Rec12-binding peaks in centromere I are found in imr1L (chr1.1605.1, see Table S1), in cnt1 (chr1.1605.3), and in imr1R (chr1.1607.2); in centromere II in cnt2 (chr2.651.2 and chr2.651.4); in centromere III in imr3L (chr3.535.1), in cnt3 (chr3.531.1 and chr3.537.2), and in imr3R (chr3.538.2).
Inverse correlation of Rec12-binding with GC content and origins of replication
Local high GC content correlates with high Spo11 binding in S. cerevisiae on a microarray with ORF probes (Gerton et al. 2000), and the third codon position GC content further increases this correlation (Birdsell 2002). We thus calculated the GC content of every ORF and intergenic probe on the microarrays. A Spearman statistical test showed a significant inverse correlation between Rec12-binding ratio and GC content in both intergenic and ORF sequences (Table 3, Fig. S3a).
Table 3.
Correlation of Rec12-binding ratios with specific chromosomal features
| Correlation of Rec12-binding with data set, applied probes on microarray (number of pairs analyzed) |
p-value | Spearman r |
|---|---|---|
| GC content, intergenic probes (6020)a |
< 0.0001 | −0.10 |
| GC content, ORF probes (4927) |
< 0.0001 | −0.16 |
| GC content, intergenic and ORF probes (10947) |
< 0.0001 | −0.15 |
| Meiotic replication origins, intergenic and ORF probes (8847)b |
< 0.0001 | −0.06 |
| Meiotic transcription, ORF probes (4685)c |
< 0.0001 | 0.09 |
| Rec8 binding, intergenic and ORF probes (5000)d |
< 0.0001 | 0.18 |
The GC content of each ORF and intergenic probe from the microarray was calculated.
Calculated with data from a pat1-114 strain treated with hydroxyurea at 3 hours into meiosis [37]
Data from the meiotic mRNA profile (pat1-114) at time point 4 hours was used [51]
Data from two Rec8 chromatin immunoprecipitation experiments (wild type) on an Affimetrix oligoarray [54] covering the length of chromosome II and part of chromosome III. The data was transformed to the intergenic and ORF array architecture (see Material and Methods).
The inverse correlation between GC content and Rec12-binding suggests that Rec12 binds preferentially in AT rich regions. AT rich islands were proposed to act as origins of replication (Segurado et al. 2003). Early replication forks in meiotic S-phase, stalled by hydroxyurea, which inhibits ribonucleotide reductase and thus depletes the nucleotide pools, were recently mapped on the same ORF and intergenic microarray platforms (Heichinger et al. 2006). A Spearman statistical test revealed a significant inverse correlation between Rec12-binding ratios and meiotic origins of replication (Table 3). Examples of this inverse correlation are shown across a 250 kb region on chromosome II in Fig. S3b.
Rec12-binding correlates with gene expression and cohesin binding
Transcription during meiosis might render chromatin in promoter regions accessible to Rec12. We therefore asked, whether genes whose transcription is induced during meiosis, were also preferred sites for Rec12-binding. Genome-wide transcription of a pat1-114 strain at 4 hours (Mata et al. 2002) was significantly correlated with the Rec12-binding ratio on the ORF sequences (Table 3). The correlation was most prominent for genes with high induction factors. Out of the 10 most strongly induced genes, 7 were also found by the CHIPOTIe algorithm as Rec12-binding peaks (C29A10.02, meu10, meu3RC, dmc1, meu4, meu19, and C22H10.13).
It was proposed for S. cerevisiae, that Spo11-binding peaks may represent regions, where initiation of recombination was not prevented by cohesin binding (Glynn et al. 2004; Mieczkowski et al. 2006). We therefore compared Rec12-binding with published data on Rec8 binding in pat1-114 haploid meiosis across chromosome II and a part of chromosome III (Ding et al. 2006). For this purpose were the Rec8-data transformed to our microarray architechture (see Material and Methods). Unexpectedly, a significantly positive correlation was evident (Table 3). A 250 kb region of chromosome II is shown in Fig. S3c. Notably, Rec12-binding peaks on chromosome II did not overlap with Rec8-binding peaks on the same probe (Table S1). Moreover, no overlap was detected for probes with a Rec12-binding ratio (log2) >1 (Table 5). But, when the limit was > 0.8, a positive correlation could be detected, suggesting that the Rec8-binding peaks might be at the edges of the Rec12-binding peaks. In fact, of 43 designated Rec12-binding peaks on chromosome II (excluding peaks in centromere II), 27 had a nearby Rec8-binding peak (Rec8-binding ratio (log2) > 0.5) within 2′500 bp on either side. In other words, 62.8% of the Rec12-binding peaks on the arms of chromosome II had a Rec8-binding peak at a distance < 2′500 bp. To test for significance, we calculated how many Rec8-binding peaks fall into random 5′000 bp windows, assuming a Rec8-binding peak width of 3′000 bp and a peak height of 0.5 (Table S6). Under this scenario, only 47.8% of the 5′000 bp windows could expect a hit, less than the 62.8% determined above. The average distance of the 27 Rec12-binding peaks to nearby Rec8-binding peaks was 1.7 kb, with a range of 55 to 2′400 bp (Fig. S4). However, due to different resolutions of the two microarrays (average probe length in Rec12-binding microarrays: 1′100 bp/probe, Rec8-binding microarray: 250 bp/probe), we cannot rule out co-localization for the smaller distances measured.
At the centromeres of chromosome II and III, co-localization of Rec12-binding and Rec8-binding was prominent. Among 20 probes of the inner repeats and the core regions (Fig. 6), we found a high correlation between Rec12- and Rec8-binding (Spearman r = 0.58, p-value < 0.0018). No correlation was found in 21 probes of the mating type region.
Discussion
Association of Rec12 with ura4A hotspot sequences
To test whether Rec12 associates with DNA sequences of a meiotic recombination hotspot, we checked Rec12 immunoprecipitated DNA from a meiotic time course by real-time PCR to ura4A sequences (Fig. 1). Experiments in a rad50+ strain showed only a slight enrichment of precipitated ura4A sequences at 4 hours (Fig. S1), which can be explained by removal of Rec12 and subsequent repair of DSBs. In rad50S mutant strains however, the amount of precipitated ura4A sequences at 4 hours was in the range of the amount of broken DNA (4-9%) at this DSB site measured by Southern blot experiments at 4 hours in haploid pat1-114 rad50S meiosis (Gregan et al. 2005). The low values at late time-points were not expected since meiotic DSBs persist in rad50S mutants. This drop of hotspot sequence recovery may be due to degradation of the Rec12 protein in the extracts. Since the immunoprecipitation was done after in vivo formaldehyde cross-linking, the values at 2 and 3 hours may indicate that Rec12 already binds before or during DNA replication, although DSB formation is not detected at these times (Gregan et al. 2005). Rec7, the S.pombe homolog of the initiation complex member Rec114 in S.cerevisiae, and the meiotic cohesin subunit Rec8 are expressed upon nitrogen starvation before the onset of DNA replication (Grishchuk 2003; Molnar et al. 2001; Parisi et al. 1999). In addition, mutants of components of the DSB formation complex advance the start of meiotic S-phase, which suggests a role before DNA synthesis (Doll 2005). Rec12myc was not proficient to precipitate ura4A hotspot sequences in the recombination deficient rec6Δ or rec14Δ mutants. There are no Rec6 sequence homologues in other organisms, but a requirement of the Sordaria macrospora Rec14 homologue, Ski8, to localize Spo11 to chromosomes in meiotic prophase was also found (Tesse et al. 2003). In S. cerevisiae however, Spo11 association with hotspot sequences was independent of the Rec14 homolog Ski8 (Prieler et al. 2005).
Rec12-binding peaks and DSB formation
We used a sliding window algorithm to identify peaks of Rec12-binding ratios from the Rec12-Chip on CHIP data, which was not otherwise filtered. The known site of strong DSB formation mbs2 (Cromie et al. 2005) was detected as a Rec12-binding peak (probe chr1.279.2, Table S1), but the mbs1 and ura4A hotspots were not among the 144 Rec12-binding peaks due to lack of data from nearby probes. However, at both sites considerable Rec12-binding was detected (2.2 for probe chr1.316.1 at mbs1, and 2.5 for probe chr3.40.1 5′ to ura4). In addition, 8 out of 11 major DSB sites on the 1′105 kb Not I fragment of chromosome I (Cromie et al. 2007) coincide with Rec12-binding ratios of more than 2, although only 2 of the CHIPOTIe Rec12-binding peaks were found among them. The criteria set for peak scoring by the ChIPOTIe algorithm were obviously rigorous. We can conclude from this accordance, that a good part of Rec12-binding comes from covalent Rec12-DNA interactions.
Rec12-binding correlates with meiotic recombination activity
Repair of meiotic DSBs can engage the sister chromatide or the homolog. If the latter is involved, repair results in a gene conversion event in the vicinity of the DSB site and/or leads to a reciprocal exchange of the involved homologs. Since Rec12 is responsible for meiotic DSBs, Rec12-binding peaks are expected to correlate with recombination activity.
No quantitative relationship between Rec12-binding ratios and gene conversion frequencies (Table 2) can be deduced from the tetrad analysis results. We neither know the location of DSBs, nor the position of the used markers, nor the steepness of conversion gradients. However, high gene conversion frequencies go along with high Rec12-binding. The marker sme2Δ::ura4 with the lowest gene conversion frequency but the highest Rec12-binding ratio (32.1) of the eight markers, is a peculiar exception. The sme2 gene encodes a meiosis-essential non-coding RNA, called meiRNA, with whom the Mei2 protein forms a dot-like structure in the nucleus at the sme2 locus during prophase (Shimada et al. 2003). High Rec12-binding with a considerably low gene conversion frequency at this site may indicate that repair of DSBs involves mostly the sister chromatid at this site. Alternatively, Rec12-binding at sme2 may serve also for another function than DSB formation (see below paragraph on centromeres).
Genetic distance on chromosome I correlates with Rec12-binding (Fig. 4). Gene conversion events without accompanying crossover (13 to 36 % depending on the locus (Cromie et al. 2005)) could explain the positive y-intercept of the linear regression line because such conversion events do not contribute to genetic distance. In addition, Rec12-binding leading to DSB repair by interaction with the sister chromatid may also contribute to the positive y-intercept. The formation of Holliday junction intermediates occurs more frequently between sister than homologous chromatids in S. pombe (Cromie et al. 2006). Such DSBs would be scored in Southern blots, but their repair would not result in crossovers or gene conversions. Overall, a good part of the Rec12-binding reflects recombination activity as observed by correlation with DSB formation, gene conversion, and crossover frequencies. But in difference to other organisms (e.g. S. cerevisiae (Gerton et al. 2000)) there are no gross differences between regions with respect to Rec12-binding and DSB formation in rad50S strains.
Correlation of strong Rec12-binding with chromosomal features
Previously, meiotic hotspots of DSB formation have been reported in large intergenic regions (Cromie et al. 2005). The high representation of Rec12-binding peaks in these regions confirms this finding. It is not obvious why the Rec12-binding pattern with respect to large gene-free regions differs for chromosome I (Fig. 5). It neither has significantly more large gene-free regions than the other chromosomes (χ2=2.5, p=0.3), nor do they significantly differ in size (see legend to Fig. 5). This finding contrasts the situation in budding yeast and could reflect differences of intergenic region lengths between the two yeast genomes (Wood et al. 2002).
Contrary to the correlation of GC content with covalent Spo11-DNA association in S. cerevisiae (Gerton et al. 2000), we find that Rec12-binding shows an inverse correlation to GC content. A similar relationship was reported in Arabidopsis thaliana where crossover rates from chromosome 4 inversely correlated with GC content (Drouaud et al. 2006). In addition, in the human MHC class II region only one out of three crossover clusters corresponded to a region of higher than average GC content (de Massy 2003). Thus, the relationship between GC content and recombination hotspots does not seem to be the same in all eukaryotes. Although replication origins and Rec12-binding sites prefer AT rich gene-free regions, they exclude each other, which would be consistent with an early loading of the DSB initiation complex before or during S-phase, as discussed above.
Promotors of genes with strong transcription during meiotic prophase are more likely to be targets of Rec12-binding. Whether this correlation is due to specific transcription-factor binding remains to be elucidated. However, it has been shown that transcription-factor binding can promote recombination activity, but is not sufficient for high recombination at all transcription factor binding sites in a given region (Kon et al. 1997; Mieczkowski et al. 2006; Steiner and Smith 2005).
Rec12-binding at centromeres
Rec12-binding peaks were detected in all three centromeric core regions (Table S1, Fig. 6). Cromie et al. showed recently a strong correlation between DSB formation and Rec12 chromatin immunoprecipitated DNA (Rec12 covalently bound to DNA, no formaldehyde cross-linking) in a rad50S mutant on a genome-wide scale (Cromie et al. 2007). They found no Rec12-DNA linkage in the centromeres. Thus, we have to assume that the detected Rec12-binding peaks in the centromere are the result of DSB-independent association. Alternatively, the absence of putative DSBs in the centromere might be an artifact of the rad50S mutant. DSBs might be resected (and repaired) through an alternative mechanism, thus escaping the rad50S trap.
In budding yeast, a role of Spo11 in early chromosome pairing and determination of S-phase length was demonstrated (Cha et al. 2000), independent of DSB and synaptonemal complex (SC) formation. Such a dual role was also proposed for Rec12 (Sharif et al. 2002). The rec12Δ mutant showed frequent non-disjunction in meiosis I. This phenotype was much milder in the rec12Y98F active site mutant, implying that Rec12 has a supporting function on chromosomes segregation in meiosis I, besides its activity in DSB formation. However, contradicting results were reported by (Davis and Smith 2003). Additional experiments are required to determine whether Rec12-dependent DSB formation occurs in centromeric core regions or whether Rec12 has another role at the centromeres.
Rec12-binding and cohesin
The correlation between Rec12-binding and Rec8-binding was strong in the centromeric core regions of chromosome II and III (Fig. 6), but was also evident for the remaining regions tested (Table 3). Partially, this correlation could be explained by the mutual preference of AT-rich sequences: a comparison of Rec8-binding and GC content revealed indeed an inverse correlation (Spearman correlation r=-0.14, p<0.0001). However, the proximity of Rec12-binding peaks to Rec8-binding peaks in more than 60% of all strong Rec12-binding peaks, let us conclude that a functional relationship between the two complexes may exist. Rec8 could direct Rec12 to susceptible regions for DSB formation. This could explain a long-known riddle: region-specific reduction of recombination in strains carrying mutations in the genes rec8, rec11, and rec10. Specific point mutations in all three genes, as well as rec8 and rec11, but not rec10 deletion strains, were found to have strongly reduced conversion and crossover frequencies (up to more than 100-fold) in the central parts of the chromosomes, but only slight (at most 10-fold) reduction at the chromosome ends (DeVeaux and Smith 1994; Li et al. 1997; Parisi et al. 1999). Regional reduction of DSB frequency in rec8Δ and rec11Δ (both genes coding for cohesin subunits) has been reported (Ellermeier and Smith 2005). Little reduction of recombination at chromosome ends may be due to the presence of mitotic cohesin complexes there during the early stages of meiosis (Kitajima et al. 2003; Yokobayashi et al. 2003). In mitosis, Rec8 and Rec11 are replaced by their paralogs Rad21 and Psc3 (Kitajima et al. 2003; Yokobayashi et al. 2003). Assistance of mitotic cohesins for the Rec12 complex would then explain the maintenance of higher recombination frequency at chromosome ends in rec8 and rec11 mutants. The presence of region-specific recombination reduction in the rec10-109 point mutant (DeVeaux and Smith 1994), but not in other rec10 point mutants (Wells et al. 2006), and complete loss of recombination in a rec10 deletion strain (Ellermeier and Smith 2005; Wells et al. 2006), might indicate that cohesin complexes and Rec12 complexes also interact with the linear element structural protein Rec10 (Lorenz et al. 2006; Molnar et al. 2003) for definition of DSB formation sites.
Supplementary Material
Fig. S1 Rec12-binding in rad50S versus rad50 wild type
Fig. S2 Location of genetic markers on chromosome I
Fig. S3 Correlation of Rec12-binding with chromosome features
Fig. S4 Distance between Rec12-binding hotspots and Rec8 binding peaks
Table S1 The 144 Rec12-binding hotspots (ChIPOTIe)
Table S2 Genetic length of different intervals on chromosome I
Table S3 Observed distribution of Rec12-binding versus random redistribution in the intervals on chromosome I
Table S4 Rec12-binding hotspots per chromosomes
Table S5 Co-localization of Rec12-binding hotspots (HS) with Rec8 binding sites on chromosome II
Table S6 Probability of occurrence of Rec8-binding peaks in Rec12-binding peak windows
Acknowledgements
We thank Hirofumi Aiba for the gpd1-E13 mutant, Andrea Ludin for writing the information retrieval programs to perform statistical analysis, Chris Penkett for help with the ArrayExpress submission, and Eva Heiniger and Silvia Steiner for construction of the rec6Δ and rec14Δ strains. We also thank Gareth Cromie and Gerry Smith, HCRC Seattle, for communication of results ahead of publication. The study was supported by the Swiss National Science Foundation to JK, and by Cancer Research U.K. [CUK], Grant No. C9546/A6517 to JB. KL was supported by a Marie-Heim-Vögtlin fellowship.
Footnotes
Supporting information: separate document “Suppl_Figures_Tables”
References
- Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Baudat F, Nicolas A. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc Natl Acad Sci U S A. 1997;94:5213–5218. doi: 10.1073/pnas.94.10.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur M, Hartsuiker E, Lehmann E, Ludin K, Munz P, Kohli J. The meiotic recombination hot spot ura4A in Schizosaccharomyces pombe. Genetics. 2005;169:551–561. doi: 10.1534/genetics.104.033647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach D, Rodgers L, Gould J. ran1+ controls the transition from mitotic division to meiosis in fission yeast. Curr Genet. 1985;10:297–311. doi: 10.1007/BF00365626. [DOI] [PubMed] [Google Scholar]
- Birdsell JA. Integrating genomics, bioinformatics, and classical genetics to study the effects of recombination on genome evolution. Mol Biol Evol. 2002;19:1181–1197. doi: 10.1093/oxfordjournals.molbev.a004176. [DOI] [PubMed] [Google Scholar]
- Borde V, Lin W, Novikov E, Petrini JH, Lichten M, Nicolas A. Association of Mre11p with double-strand break sites during yeast meiosis. Mol Cell. 2004;13:389–401. doi: 10.1016/s1097-2765(04)00034-6. [DOI] [PubMed] [Google Scholar]
- Buck MJ, Nobel AB, Lieb JD. ChIPOTle: a user-friendly tool for the analysis of ChIP-chip data. Genome Biol. 2005;6:R97. doi: 10.1186/gb-2005-6-11-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes MD, Farah JA, Smith GR. Meiotic DNA breaks associated with recombination in S. pombe. Mol Cell. 2000;5:883–888. doi: 10.1016/s1097-2765(00)80328-7. [DOI] [PubMed] [Google Scholar]
- Cha RS, Weiner BM, Keeney S, Dekker J, Kleckner N. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev. 2000;14:493–503. [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Rubio CA, Hyppa RW, Smith GR. A Natural Meiotic DNA Break Site in Schizosaccharomyces pombe Is a Hotspot of Gene Conversion, Highly Associated With Crossing Over. Genetics. 2005;169:595–605. doi: 10.1534/genetics.104.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Hyppa RW, Cam HP, Farah JA, Grewal SI, Smith GR. A discrete class of intergenic DNA dictates meiotic DNA break hotspots in fission yeast. PLoS Genet. 2007;3:e141. doi: 10.1371/journal.pgen.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, Smith GR. Nonrandom homolog segregation at meiosis I in Schizosaccharomyces pombe mutants lacking recombination. Genetics. 2003;163:857–874. doi: 10.1093/genetics/163.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B. Distribution of meiotic recombination sites. Trends Genet. 2003;19:514–522. doi: 10.1016/S0168-9525(03)00201-4. [DOI] [PubMed] [Google Scholar]
- DeVeaux LC, Smith GR. Region-specific activators of meiotic recombination in Schizosaccharomyces pombe. Genes Dev. 1994;8:203–210. doi: 10.1101/gad.8.2.203. [DOI] [PubMed] [Google Scholar]
- Ding DQ, Sakurai N, Katou Y, Itoh T, Shirahige K, Haraguchi T, Hiraoka Y. Meiotic cohesins modulate chromosome compaction during meiotic prophase in fission yeast. J Cell Biol. 2006;174:499–508. doi: 10.1083/jcb.200605074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll E. PhD thesis. University of Bern; 2005. [Google Scholar]
- Drouaud J, Camilleri C, Bourguignon PY, Canaguier A, Berard A, Vezon D, Giancola S, Brunel D, Colot V, Prum B, Quesneville H, Mezard C. Variation in crossing-over rates across chromosome 4 of Arabidopsis thaliana reveals the presence of meiotic recombination “hot spots”. Genome Res. 2006;16:106–114. doi: 10.1101/gr.4319006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier C, Smith GR. Cohesins are required for meiotic DNA breakage and recombination in Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 2005;102:10952–10957. doi: 10.1073/pnas.0504805102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DH, Li YF, Fox ME, Smith GR. A WD repeat protein, Rec14, essential for meiotic recombination in Schizosaccharomyces pombe. Genetics. 1997;146:1253–1264. doi: 10.1093/genetics/146.4.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton JL, DeRisi J, Shroff R, Lichten M, Brown PO, Petes TD. Inaugural article: global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2000;97:11383–11390. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregan J, Rabitsch PK, Sakem B, Csutak O, Latypov V, Lehmann E, Kohli J, Nasmyth K. Novel genes required for meiotic chromosome segregation are identified by a high-throughput knockout screen in fission yeast. Curr Biol. 2005;15:1663–1669. doi: 10.1016/j.cub.2005.07.059. [DOI] [PubMed] [Google Scholar]
- Grimm C, Kohli J, Murray J, Maundrell K. Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 gene as a selectable marker. Mol Gen Genet. 1988;215:81–86. doi: 10.1007/BF00331307. [DOI] [PubMed] [Google Scholar]
- Grishchuk AL. PhD thesis. University of Bern; Switzerland: 2003. [Google Scholar]
- Gutz H. Site specific induction of gene conversion in Schizosaccharomyces pombe. Genetics. 1971;69:317–337. doi: 10.1093/genetics/69.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygax AT, P. A revised chromosome map of the fission yeast Schizosaccharomyces pombe. Curr Genet. 1984;8:85–92. doi: 10.1007/BF00420223. [DOI] [PubMed] [Google Scholar]
- Heichinger C, Penkett CJ, Bähler J, Nurse P. Genome-wide characterization of fission yeast DNA replication origins. Embo J. 2006;25:5171–5179. doi: 10.1038/sj.emboj.7601390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentges P, Van Driessche B, Tafforeau L, Vandenhaute J, Carr AM. Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast. 2005;22:1013–1019. doi: 10.1002/yea.1291. [DOI] [PubMed] [Google Scholar]
- Keeney S. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick DT, Wang YH, Dominska M, Griffith JD, Petes TD. Control of meiotic recombination and gene expression in yeast by a simple repetitive DNA sequence that excludes nucleosomes. Mol Cell Biol. 1999;19:7661–7671. doi: 10.1128/mcb.19.11.7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima TS, Yokobayashi S, Yamamoto M, Watanabe Y. Distinct cohesin complexes organize meiotic chromosome domains. Science. 2003;300:1152–1155. doi: 10.1126/science.1083634. [DOI] [PubMed] [Google Scholar]
- Kon N, Krawchuk MD, Warren BG, Smith GR, Wahls WP. Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1997;94:13765–13770. doi: 10.1073/pnas.94.25.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- Lambie EJ, Roeder GS. Repression of meiotic crossing over by a centromere (CEN3) in Saccharomyces cerevisiae. Genetics. 1986;114:769–789. doi: 10.1093/genetics/114.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambie EJ, Roeder GS. A yeast centromere acts in cis to inhibit meiotic gene conversion of adjacent sequences. Cell. 1988;52:863–873. doi: 10.1016/0092-8674(88)90428-x. [DOI] [PubMed] [Google Scholar]
- Li YF, Numata M, Wahls WP, Smith GR. Region-specific meiotic recombination in Schizosaccharomyces pombe: the rec11 gene. Mol Microbiol. 1997;23:869–878. doi: 10.1046/j.1365-2958.1997.2691632.x. [DOI] [PubMed] [Google Scholar]
- Lobry JR. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- Lorenz A, Estreicher A, Kohli J, Loidl J. Meiotic recombination proteins localize to linear elements in Schizosaccharomyces pombe. Chromosoma. 2006 doi: 10.1007/s00412-006-0053-9. [DOI] [PubMed] [Google Scholar]
- Lyne R, Burns G, Mata J, Penkett CJ, Rustici G, Chen D, Langford C, Vetrie D, Bähler J. Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics. 2003;4:27. doi: 10.1186/1471-2164-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleki S, Neale MJ, Arora C, Henderson KA, Keeney S. Interactions between Mei4, Rec114, and other proteins required for meiotic DNA double-strand break formation in Saccharomyces cerevisiae. Chromosoma. 2007 doi: 10.1007/s00412-007-0111-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Lyne R, Burns G, Bähler J. The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet. 2002;32:143–147. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- Mieczkowski PA, Dominska M, Buck MJ, Gerton JL, Lieb JD, Petes TD. Global analysis of the relationship between the binding of the Bas1p transcription factor and meiosis-specific double-strand DNA breaks in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:1014–1027. doi: 10.1128/MCB.26.3.1014-1027.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrenweiser HW, Tsujimoto S, Gordon L, Olsen AS. Regions of sex-specific hypo- and hyper-recombination identified through integration of 180 genetic markers into the metric physical map of human chromosome 19. Genomics. 1998;47:153–162. doi: 10.1006/geno.1997.5097. [DOI] [PubMed] [Google Scholar]
- Molnar M, Parisi S, Kakihara Y, Nojima H, Yamamoto A, Hiraoka Y, Bozsik A, Sipiczki M, Kohli J. Characterization of rec7, an early meiotic recombination gene in Schizosaccharomyces pombe. Genetics. 2001;157:519–532. doi: 10.1093/genetics/157.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar M, Doll E, Yamamoto A, Hiraoka Y, Kohli J. Linear element formation and their role in meiotic sister chromatid cohesion and chromosome pairing. J Cell Sci. 2003;116:1719–1731. doi: 10.1242/jcs.00387. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Munz P. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics. 1994;137:701–707. doi: 10.1093/genetics/137.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko Y, Adachi Y, Funahashi S, Niwa O, Yanagida M. Chromosome walking shows a highly homologous repetitive sequence present in all the centromere regions of fission yeast. Embo J. 1986;5:1011–1021. doi: 10.1002/j.1460-2075.1986.tb04316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas A, Treco D, Schultes NP, Szostak JW. An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature. 1989;338:35–39. doi: 10.1038/338035a0. [DOI] [PubMed] [Google Scholar]
- Nurse P. Genetic control of cell size at cell division in yeast. Nature. 1975;256:547–551. doi: 10.1038/256547a0. [DOI] [PubMed] [Google Scholar]
- Ohmiya R, Yamada H, Nakashima K, Aiba H, Mizuno T. Osmoregulation of fission yeast: cloning of two distinct genes encoding glycerol-3-phosphate dehydrogenase, one of which is responsible for osmotolerance for growth. Mol Microbiol. 1995;18:963–973. doi: 10.1111/j.1365-2958.1995.18050963.x. [DOI] [PubMed] [Google Scholar]
- Parisi S, McKay MJ, Molnar M, Thompson MA, van der Spek PJ, van Drunen-Schoenmaker E, Kanaar R, Lehmann E, Hoeijmakers JH, Kohli J. Rec8p, a meiotic recombination and sister chromatid cohesion phosphoprotein of the Rad21p family conserved from fission yeast to humans. Mol Cell Biol. 1999;19:3515–3528. doi: 10.1128/mcb.19.5.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkett CJ, Birtle ZE, Bahler J. Simplified primer design for PCR-based gene targeting and microarray primer database: two web tools for fission yeast. Yeast. 2006;23:921–928. doi: 10.1002/yea.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieler S, Penkner A, Borde V, Klein F. The control of Spo11’s interaction with meiotic recombination hotspots. Genes Dev. 2005;19:255–269. doi: 10.1101/gad.321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakem B. PhD thesis. University of Bern; 2005. [Google Scholar]
- Segurado M, de Luis A, Antequera F. Genome-wide distribution of DNA replication origins at A+T-rich islands in Schizosaccharomyces pombe. EMBO Rep. 2003;4:1048–1053. doi: 10.1038/sj.embor.7400008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif WD, Glick GG, Davidson MK, Wahls WP. Distinct functions of S. pombe Rec12 (Spo11) protein and Rec12-dependent crossover recombination (chiasmata) in meiosis I; and a requirement for Rec12 in meiosis II. Cell Chromosome. 2002;1:1. doi: 10.1186/1475-9268-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Yamashita A, Yamamoto M. The fission yeast meiotic regulator Mei2p forms a dot structure in the horse-tail nucleus in association with the sme2 locus on chromosome II. Mol Biol Cell. 2003;14:2461–2469. doi: 10.1091/mbc.E02-11-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner WW, Schreckhise RW, Smith GR. Meiotic DNA breaks at the S. pombe recombination hot spot M26. Mol Cell. 2002;9:847–855. doi: 10.1016/s1097-2765(02)00489-6. [DOI] [PubMed] [Google Scholar]
- Steiner WW, Smith GR. Natural meiotic recombination hot spots in the Schizosaccharomyces pombe genome successfully predicted from the simple sequence motif M26. Mol Cell Biol. 2005;25:9054–9062. doi: 10.1128/MCB.25.20.9054-9062.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesse S, Storlazzi A, Kleckner N, Gargano S, Zickler D. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc Natl Acad Sci U S A. 2003;100:12865–12870. doi: 10.1073/pnas.2034282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Miyashita K, Saito TT, Nabeshima K, Nojima H. Abundant poly(A)-bearing RNAs that lack open reading frames in Schizosaccharomyces pombe. DNA Res. 2002;9:209–215. doi: 10.1093/dnares/9.6.209. [DOI] [PubMed] [Google Scholar]
- Wells JL, Pryce DW, Estreicher A, Loidl J, McFarlane RJ. Linear element-independent meiotic recombination in Schizosaccharomyces pombe. Genetics. 2006;174:1105–1114. doi: 10.1534/genetics.106.063818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winckler W, Myers SR, Richter DJ, Onofrio RC, McDonald GJ, Bontrop RE, McVean GA, Gabriel SB, Reich D, Donnelly P, Altshuler D. Comparison of fine-scale recombination rates in humans and chimpanzees. Science. 2005;308:107–111. doi: 10.1126/science.1105322. [DOI] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, Bowman S, Brooks K, Brown D, Brown S, Chillingworth T, Churcher C, Collins M, Connor R, Cronin A, Davis P, Feltwell T, Fraser A, Gentles S, Goble A, Hamlin N, Harris D, Hidalgo J, Hodgson G, Holroyd S, Hornsby T, Howarth S, Huckle EJ, Hunt S, Jagels K, James K, Jones L, Jones M, Leather S, McDonald S, McLean J, Mooney P, Moule S, Mungall K, Murphy L, Niblett D, Odell C, Oliver K, O’Neil S, Pearson D, Quail MA, Rabbinowitsch E, Rutherford K, Rutter S, Saunders D, Seeger K, Sharp S, Skelton J, Simmonds M, Squares R, Squares S, Stevens K, Taylor K, Taylor RG, Tivey A, Walsh S, Warren T, Whitehead S, Woodward J, Volckaert G, Aert R, Robben J, Grymonprez B, Weltjens I, Vanstreels E, Rieger M, Schafer M, Muller-Auer S, Gabel C, Fuchs M, Dusterhoft A, Fritzc C, Holzer E, Moestl D, Hilbert H, Borzym K, Langer I, Beck A, Lehrach H, Reinhardt R, Pohl TM, Eger P, Zimmermann W, Wedler H, Wambutt R, Purnelle B, Goffeau A, Cadieu E, Dreano S, Gloux S, Lelaure V, Mottier S, Galibert F, Aves SJ, Xiang Z, Hunt C, Moore K, Hurst SM, Lucas M, Rochet M, Gaillardin C, Tallada VA, Garzon A, Thode G, Daga RR, Cruzado L, Jimenez J, Sanchez M, del Rey F, Benito J, Dominguez A, Revuelta JL, Moreno S, Armstrong J, Forsburg SL, Cerutti L, Lowe T, McCombie WR, Paulsen I, Potashkin J, Shpakovski GV, Ussery D, Barrell BG, Nurse P. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Xu F, Petes TD. Fine-structure mapping of meiosis-specific double-strand DNA breaks at a recombination hotspot associated with an insertion of telomeric sequences upstream of the HIS4 locus in yeast. Genetics. 1996;143:1115–1125. doi: 10.1093/genetics/143.3.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Mizuno KI, Hirota K, Kon N, Wahls WP, Hartsuiker E, Murofushi H, Shibata T, Ohta K. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. Embo J. 2004;23:1792–1803. doi: 10.1038/sj.emboj.7600138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Hiraoka Y. Monopolar spindle attachment of sister chromatids is ensured by two distinct mechanisms at the first meiotic division in fission yeast. Embo J. 2003;22:2284–2296. doi: 10.1093/emboj/cdg222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobayashi S, Yamamoto M, Watanabe Y. Cohesins determine the attachment manner of kinetochores to spindle microtubules at meiosis I in fission yeast. Mol Cell Biol. 2003;23:3965–3973. doi: 10.1128/MCB.23.11.3965-3973.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JA, Schreckhise RW, Steiner WW, Smith GR. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol Cell. 2002;9:253–263. doi: 10.1016/s1097-2765(02)00452-5. [DOI] [PubMed] [Google Scholar]
- Yu A, Zhao C, Fan Y, Jang W, Mungall AJ, Deloukas P, Olsen A, Doggett NA, Ghebranious N, Broman KW, Weber JL. Comparison of human genetic and sequence-based physical maps. Nature. 2001;409:951–953. doi: 10.1038/35057185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Rec12-binding in rad50S versus rad50 wild type
Fig. S2 Location of genetic markers on chromosome I
Fig. S3 Correlation of Rec12-binding with chromosome features
Fig. S4 Distance between Rec12-binding hotspots and Rec8 binding peaks
Table S1 The 144 Rec12-binding hotspots (ChIPOTIe)
Table S2 Genetic length of different intervals on chromosome I
Table S3 Observed distribution of Rec12-binding versus random redistribution in the intervals on chromosome I
Table S4 Rec12-binding hotspots per chromosomes
Table S5 Co-localization of Rec12-binding hotspots (HS) with Rec8 binding sites on chromosome II
Table S6 Probability of occurrence of Rec8-binding peaks in Rec12-binding peak windows