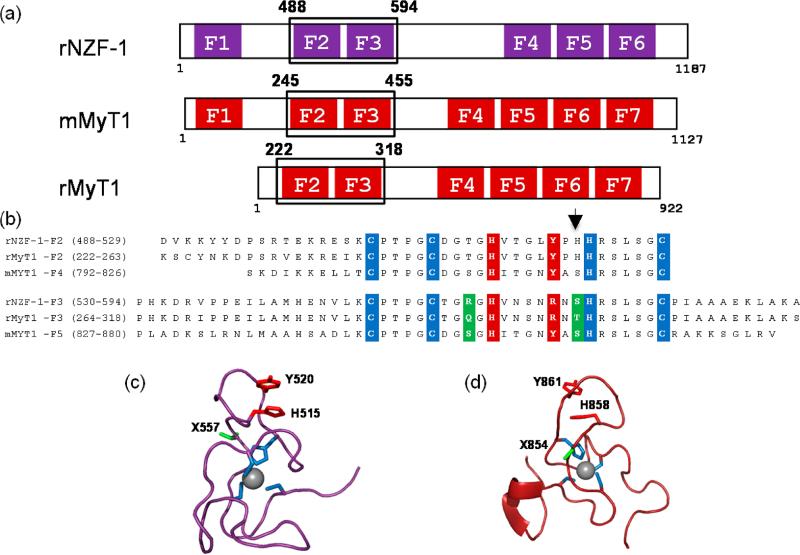
Figure 1.
(a) Cartoon diagram of the ZF topology of NZF-1 and MyT1 from Rattus norvegicus, r or Mus musculus, m. The individual ZF domains are boxed, and the alignment shows the ZF clustering. Note, F1 is absent in rMyT1. (b) Alignment of the amino acid sequences of NZF-1-F2F3, MyT1-F2F3, and MyT1-F4F5. The cysteine and histidine ligands that can directly coordinate Co(II) and Zn(II) are highlighted in blue, the amino acid residues that have been proposed to participate in a stacking interaction with the noncoordinating histidine are highlighted in red, and the amino acid residues that differ between the individual ZF domains of NZF-1 and MyT1 are highlighted in green. The arrow indicates the position of the nonconserved histidine residue. (c) The NMR solution structure of F2 of rNZF-1 (PDBID IPXE). The highlighted amino acids are color coded to match those that are highlighted in panel 1b. The amino acid position found to be important for NZF-1-F2F3 DNA recognition is indicated in green (X557). (d) NMR solution structure of finger 5 mMyT1 (PDBID 2JYD). Highlighted amino acids are color coded to match those in panel 1b. The amino acid position that has been shown to be important for NZF-1-F2F3 DNA recognition are highlighted in green. The structural figures were generated in Pymol.