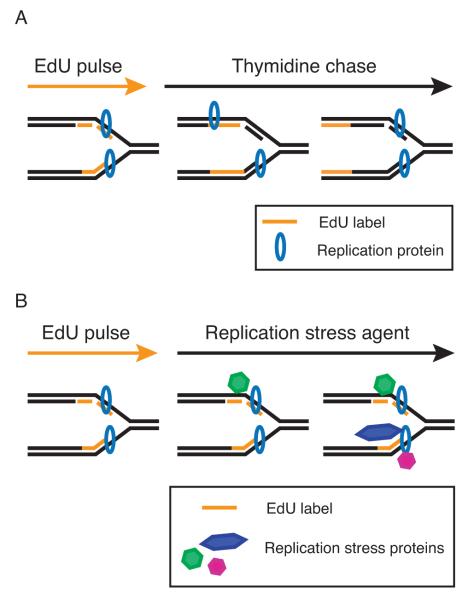
Fig. 3. Schematic of experimental procedures used to identify replisome or DNA damage proteins and modifications at the replication fork.
A. To identify replisome proteins, a pulse-chase variation of the iPOND protocol employs a thymidine chase to move the nascent, EdU-labeled DNA segment away from the replication fork. The chase sample provides a control to distinguish replisome components from general chromatin binding factors. B. To study proteins and modifications associated with damaged replication forks, an agent that stalls replication forks such as hydroxyurea is added following the EdU labeling period.