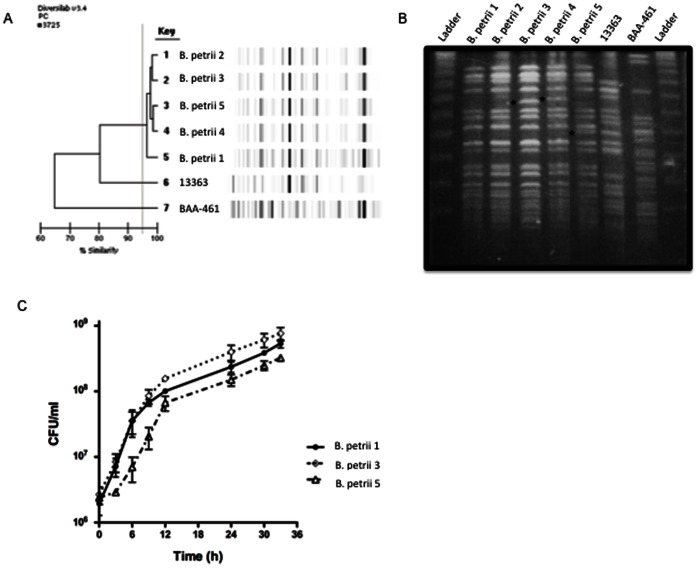
Figure 1. Molecular typing and growth curves of serially isolated strains of B. petrii..
(A) The DiversiLab Non fermentor typing kit was used for rep-PCR typing of B. petrii using DNA from clinical and reference strains. Amplicons were detected with the Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA) and data analyzed with the DiversiLab software (version 3.3). Results generated include a dendrogram (left) and virtual gel images (right). (B) Genomic DNA was digested with the restriction endonuclease XbaI and separated by PFGE with a CHEF Mapper system. Asterisks indicate band differences among the patient strains. Ladder: Lambda DNA Ladder 48.5 KB–1 MB kb plugs (Lonza). (C) Growth curves were performed on LB broth at 37°C and growth assessed by colony-forming units (CFU) performed with serially diluted aliquots plated on SBA plates. Graph shows mean and SEM from three experiments. B. petrii 1–5: strains of B. petrii serially isolated from our patient; BAA-461: type strain of B. petrii (ATCC BAA-461); 13363: first described clinical strain of B. petrii (NCTC 13363).